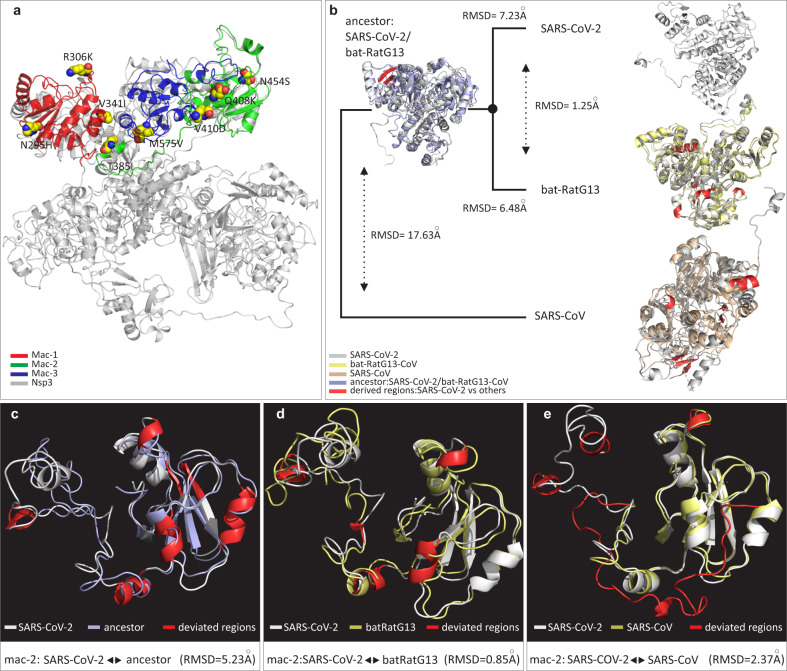
Fig. 3. Protein structural analysis of macrodomains examining the effects of specific fixed substitutions within Nsp3 of SARS-CoV-2.
a Protein structure of SARS-CoV-2 Nsp3 depicting types and locations of fixed amino-acid substitutions within distinct macrodomains. The fixed amino-acid replacements are shown as spheres and labeled with the amino-acid position in Nsp3 protein. b Comparison of the three-dimensional (3D) conformations of the macrodomains (Mac-1, Mac-2, and Mac-3) within Nsp3. SARS-CoV-2 (YP_009725299.1) macrodomains were also compared to the corresponding homologous protein regions of bat-RatG13 (QHR63299.1), SARS-CoVBJ01 (AAP30028.1), and the predicted ancestral macrodomain of SARS-CoV-2 and bat-RaTG13/ZC45/ZXC21. Structural deviations in terms of backbone torsion angles (Φ°, Ψ°) are represented in red color and were examined by RMSD (root mean square deviation) values. c–e Close-up of 3D conformations of SARS-CoV-2 Mac-2 with corresponding homologous domains from predicted ancestor (aforementioned; left panel), bat-RatG13 (middle panel), and SARS-CoV (AAP30028.1; right panel). Comparisons of 3D conformations for Mac-1 is provided in Fig. 4 and sequence secondary structural level details for comparisons in b–e are given in Supplementary Tables S6, S7.