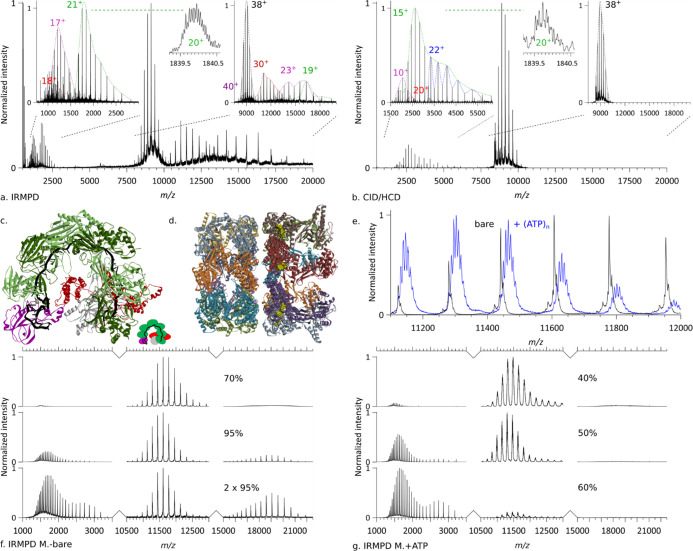
Figure 6.
(a–c) IRMPD and CID/HCD of SCRI104 CRISPR–Cas Csy complex, see Table S2 for additional information. (a) IRMPD of (black envelope, 38+) precursor (pr.) leading to the ejection of Csy3 (green, 21+) with an isotopically resolved peak (inset, 20+) and Cas6f (purple, 17+), as well as a subunit fragment (red, 18+), and the formation of pr.–Cas6f (purple, 40+), pr.–fragment (red, 30+), pr.–Cas6f (purple, 23+), pr.–Csy3 (green, 19+). Collision energy = 0 direct eV, 1.44 J/pulse, p(N2) = 1.29 × 10–10 mbar, resolution 140k @ m/z 200, no micro-scan averaging. (b) CID/HCD of (black envelope, 38+) precursor leading to the ejection of Csy3 dimer (blue, 22+), Csy1 (red, 20+), Csy3 (green, 15+) with zoom-on isotopic distribution (inset, 20+), and Cas6f (purple, 10+). Collision energy = 120 direct eV, no laser, p(N2) = 3.22 × 10–10 mbar, resolution 140k @ m/z 200, 10 micro-scans averaging. (c) Structural model of an analogous CRISPR–Cas complex (PDB ID: 5UZ9). (d–g) IRMPD of GroEL loaded with ATP. (d) Structural model of GroEL (PDB ID: 4AAS). (e) Comparison of the mass spectra of (black) bare GroEL and (blue) GroEL loaded with ATP. (f) IRMPD of bare GroEL for different irradiances and laser shot numbers. Collision energy = 1 direct eV, p(N2) = 1.27 × 10–10 mbar, resolution 140k @ m/z 200, no microscans averaging. (c) Structural model of an analogous CRISPR–Cas complex (PDB ID: 5UZ9). (g) IRMPD of GroEL loaded with ATP at different irradiances. Collision energy = 1 direct eV, p(N2) = 1.61 × 10–10 mbar, resolution 140k @ m/z 200, no micro-scans averaging.