Abstract
The development of Next-Generation Sequencing (NGS) has provided useful diagnostic, prognostic, and therapeutic strategies for individualized management of B-cell precursor acute lymphoblastic leukemia (BCP-ALL) patients. Consequently, NGS is rapidly being established in clinical practice. However, the technology’s complexity, bioinformatics analysis, and the different available options difficult a broad consensus between different laboratories in its daily routine introduction. This collaborative study among Spanish centers was aimed to assess the feasibility, pros, and cons of our customized panel and other commercial alternatives of NGS-targeted approaches. The custom panel was tested in three different sequencing centers. We used the same samples to assess other commercial panels (OncomineTM Childhood Cancer Research Assay; Archer®FusionPlex® ALL, and Human Comprehensive Cancer Panel GeneRead Panel v2®). Overall, the panels showed a good performance in different centers and platforms, but each NGS approach presented some issues, as well as pros and cons. Moreover, a previous consensus on the analysis and reporting following international guidelines would be preferable to improve the concordance in results among centers. Our study shows the challenges posed by NGS methodology and the need to consider several aspects of the chosen NGS-targeted approach and reach a consensus before implementing it in daily practice.
Keywords: next-generation sequencing, NGS-targeted panel, childhood acute lymphoblastic leukemia
1. Introduction
Acute lymphoblastic leukemia (ALL) is the most common pediatric cancer and the primary cause of death related to cancer in childhood [1,2,3]. Pediatric ALL’s survival rates have improved significantly due to better prognostic stratification and risk-directed therapies [4]. However, a relevant proportion of patients relapse, and short and long-term side effects are the main concern in survivors [5]. Thus, more personalized medicine is still needed. The recent advent of Next-Generation Sequencing (NGS) technologies has provided the methodology to find useful biomarkers to tailor treatment and manage patients individually. NGS provides the potential for increased throughput, sensitivity, and quantification of variant allelic frequencies (VAF) compared to Sanger sequencing [6]. Importantly, each NGS step can influence the results: DNA extraction, library preparation, sequencing technologies, quality controls, bioinformatic filtering, biological interpretation in the clinical context, and reporting. Moreover, the existence of several NGS platforms allows the comparison of different approaches but also makes standardization indispensable to provide homogeneous and reliable results. Global analyses of the entire genome (Whole-Genome Sequencing, WGS) or exome (Whole-Exome Sequencing, WES) are available in research. Still, few of these approaches are offered by clinical laboratories to guide routine clinical practice: they remain expensive, time-consuming, and laborious [7]. Therefore, we need to incorporate easier and cost-effective targeted sequencing approaches into clinical practice [8]. Targeted approaches require design, library preparation protocols, and enrichment assays to achieve accurate variant calling that enables us to define relevant causative variants, but they offer better base-pair coverage, running times, costs, and easier dataset handling than WGS and WES [9]. Optimal panels should provide useful information on diagnosis, prognosis, and potential therapeutic targets and remain easy to use in daily routine settings. Nowadays, there are a plethora of different custom-NGS panels developed by research groups and commercially available panels. Thus, although several options for genomic patients profiling are currently available, it is important to perform a detailed study of their scope before choosing them. Finally, the appropriate analysis, interpretation of the pathogenicity, and integration with other molecular findings remain a major challenge, especially for novel variants, those of uncertain significance, as well as for incidental findings [10].
This study is a collaborative project of the Biological Committee of the Group of Leukemia of the Spanish Society of Pediatric Hematology and Oncology (SEHOP). We aimed to test four NGS panels (one custom panel and three commercial panels) for their application in clinical routine, in the setting of the diagnosis of B-cell precursor ALL (BCP-ALL). We dissect each panel and discuss their design, analysis, and applicability related to BCP-ALL. Our study also examines the challenges that lay ahead before introducing and harmonizing the NGS-targeted approaches in clinical routine for childhood BCP-ALL diagnosis. The present study may offer helpful criteria for hematological genetic laboratories when implementing new NGS panels.
2. Materials and Methods
2.1. Ethical Issues
According to the national and international guidelines, the study strictly followed the ethical standards and the Declaration of Helsinki. All the samples were stored in the Biobank of Hospital Sant Joan de Déu (HSJD) and, according to our Ethics Committee. Samples were used after written informed consent was obtained from the patients or their legal guardians.
2.2. Design of the Study
The current study aimed to test on a pilot cohort of pediatric BCP-ALL patients, the pros and cons of each panel, and highlight critical points in NGS panel choice to improve BCP-ALL diagnostics. The following factors were considered: (1) the same assay was tested in different centers; (2) the same samples were tested on various assays; (3) to reproduce real-life faithfully, no previous consensus in bioinformatics pipelines and reporting was achieved before the study.
Thus, four NGS-targeted panels were tested: (1) the BCP-ALL custom panel; (2) Archer® FusionPlex® ALL kit (ArcherDX, Boulder, CO, USA); (3) OncomineTM Childhood Cancer Research Assay (OCCRA) (ThermoFisher Scientific, Waltham, MA, USA); and (4) Human Comprehensive Cancer GeneRead Panel v2® (Qiagen, Hilden, Germany), using Illumina (San Diego, CA, USA) and Ion Torrent (Gilford, NH, USA) platforms.
To assess the performance of the different panels, DNA and RNA from 4 BCP-ALL pediatric patients (see Supplementary Material for detailed information) were distributed. Those samples have been previously diagnosed and genetically characterized in center 1 (HSJD) by conventional methods. To evaluate the harmonization between different centers in NGS testing implementation, the samples were sent to center 2: Hospital Universitario de Salamanca (HUS); center 3: Hospital Niño Jesús (HNJ)-Instituto de Salud Carlos III (ISCIII); and center 4: Hospital La Fe (HLF). The main characteristics of patients are detailed in Supplementary Table S1.
The workflow of samples, panels, and platforms is shown in Supplementary Figure S1.
2.3. NGS-Targeted Panels Included in the Study
In this study, we have examined the performance of four NGS-targeted panels. To reproduce real-life faithfully in clinical laboratories, the panel selection was made following different criteria in individual centers, including NGS panel target design, equipment, and human and economic resources. We started our study by dissecting the currently available information about panel specifications, such as technical specifications and genes included (see below).
The genes present in the four panels are listed in Supplementary Tables S2–S5.
2.3.1. BCP-ALL Custom Panel
The BCP-ALL custom panel was designed to screen for single nucleotide variants (SNVs) and copy number variant (CNV). We selected 69 genes of diagnostic, prognostic, or therapeutic interest recurrently identified in BCP-ALL according to the literature. Details on the probe design are provided in Supplementary Methods.
2.3.2. Archer® FusionPlex® ALL Kit
The Archer® FusionPlex® ALL Kit uses 506 gene-specific primers (GSPs) that target 81 genes commonly mutated in ALL. This panel uses the Anchored Multiplex PCR (AMP), which allows identifying fusions with both known and unknown partners. For the library generation, two different protocols were applied following the manufacturer’s specification depending on the sequencing platform used (Illumina or Ion Torrent). Details on the performance are provided in Supplementary Methods.
2.3.3. OncomineTM Childhood Cancer Research Assay (OCCRA)
The OncomineTM Childhood Cancer Research Assay (Ion Torrent, ThermoFisher) is a pan-pediatric cancer panel. It includes the analysis in hot spots regions of 82 genes, the full coding sequence of 44 targets, and the CNV variation in 24 genes. It can also detect rearrangements of 88 drivers and quantify the expression of 9 targets when RNA is analyzed. Details on the performance are provided in Supplementary Methods.
2.3.4. Human Comprehensive Cancer GeneRead Panel v2®
This commercial panel includes 160 genes frequently mutated in cancer. Details on the performance are provided in Supplementary Methods.
2.4. Sequencing and Variant Data Analysis
Sequencing was performed using Illumina (San Diego, CA, USA) and Ion Torrent (Gilford, NH, USA) platforms depending on the panel manufacturer’s specifications. Further details on sequencing and bioinformatics pipelines are provided in Supplementary Methods.
The main criteria for manual variant prioritization in center 1 were: (1) exclusion of variants with a population frequency (Minor Allele Frequency, MAF) of ≥1% in the 1000 Genomes database; (2) exclusion of synonymous variants; (3) inclusion of only exonic variants (intronic regions were set aside for a second reanalysis); and (4) calls with a VAF ≥ 5% and minimum coverage of 50× per variant, which were considered to be of enough quality to be interpreted and potentially reported. The Integrative Genomics Viewer (IGV) (Broad Institute and the Regents of the University of California) version 2.5.011 was used to visually assess the authenticity of individual calls and exclude potential artifacts. To prioritize variants in centers 2, 3, and 4, synonymous variants, noncoding variants, or variants present in population databases (including dbSNP144, the 1000 genomes Project, ExAC, ESP 6500, and in-house database) at a MAF ≥ 1% were discarded. Variants recurrently observed and suspected of being sequencing errors by visual inspection on the IGV version 2.3.68 were removed. However, all the known hotspot mutations and the variants described in the Catalogue of Somatic Mutations in Cancer database (COSMIC82 database) and/or in ClinVar were rescued. To elucidate the effects of variants with no clear clinical significance, the PolyPhen-2, SIFT, and Mutation Taster web-based platforms were used. The mutations with deleterious and/or probably damaging effects were also considered as somatic mutations, whereas the remaining variants were considered candidate somatic mutations.
For CNV detection, the mean coverage depth of an individual exon target of a sample was first normalized to the total amount of the DNA template loaded onto sequencing flow cells, based on this sample’s total reads. The mean coverage of each target from the reference samples obtained was used as the reference for a specific exon. A reference coverage profile for a normal sample was generated. Finally, the normalized coverage of each exon of a test sample was compared to the mean coverage of the same target in the reference file generated above [9].
3. Results
The four samples were sequenced using the custom panel and three commercially available NGS panels. Results from DNA sequencing are shown in Supplementary Table S6.
The previously characterized mutations with the custom panel were found with the commercial panels if the design covered their genomic location.
3.1. Dissection of the NGS Panels
3.1.1. BCP-ALL Custom Panel
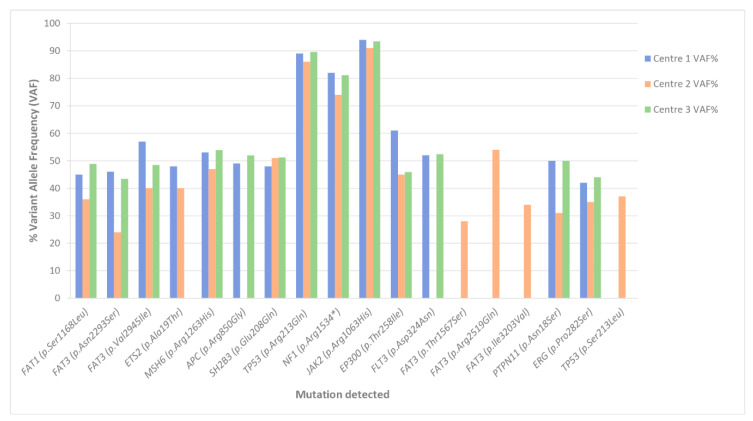
Our BCP-ALL custom panel was intended to search for SNVs and CNVs by using DNA as a template. All samples passed the quality controls. The final molarity loaded on the NextSeq flow cell was 1.6 pM. NextSeq cluster densities achieved for the BCP-ALL custom panel libraries were as expected, with a median of 5.67 million reads and 93.6% on-target coverage. To assess the reproducibility, the four samples were sequenced independently in three different centers. Sequencing metrics, variant detection, and VAF were highly reproducible between centers, and the results were in concordance with those obtained by other methodologies (Figure 1 and Supplementary Table S6). We compared all variants obtained before data curation, so we included in the comparison all the different variants detected, including Single Nucleotide Polymorphism (SNPs).
Figure 1.
Variant allele frequency of variants detected by each center in the four common samples analyzed, using the B-cell precursor acute lymphoblastic leukemia (BCP-ALL) custom panel.
Overall, the BCP-ALL custom panel detected a mean of 0.75 variations per patient in 13 genes. All were SNVs (n = 3), and neither small frameshift insertions nor deletions were found.
Those variants with VAF around 50% were studied in matched samples obtained in complete remission (CR) with negative minimal residual disease (MRD) (and thus considered non-somatic): FAT1, FAT3, and ETS2 in sample 1; MSH6, APC, and SH2B3 in sample 2; EP300, JAK2, FLT3, and FAT3 in sample 3, and PTPN11 and ERG in sample 4. Overall, only TP53 and NF1 in sample 2 were confirmed as somatic mutations and were detected by all centers. Furthermore, NF1 variant was detected in homozygosis; this fact is probably related to the loss of genetic material evidenced in the karyotype (see Supplementary Table S1).
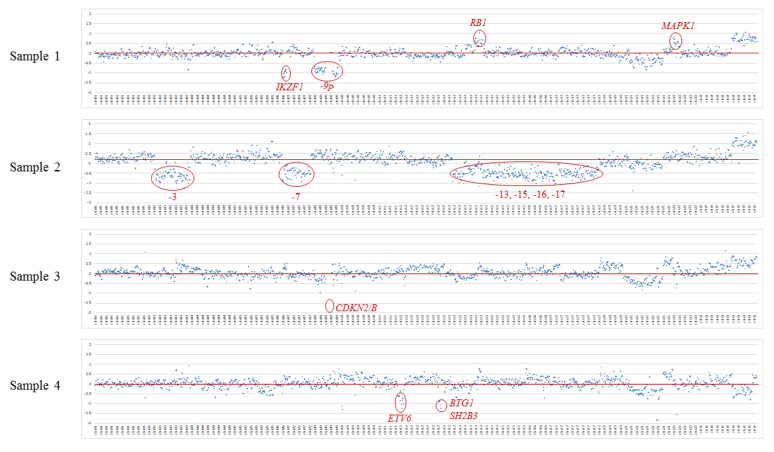
CNV analysis confirmed the presence of alterations previously identified by karyotype and/or Multiplex Ligation-Dependent Probe Amplification (MLPA). Sample 2 showed the loss of chromosomes 3, 7, 13, 15, 16, and 17, consistent with the corresponding monosomies and the low hypodiploidy observed in the karyotype and the pattern obtained by the MLPA, P181, and P182 kits. Additionally, the custom panel detected a focal deletion of IKZF1, loss of 9p, and gains of RB1 and MAPK1 in sample 1, loss of CDKN2A/B in sample 3, and loss of ETV6, BTG1, and SH2B3 genes in sample 4 (Figure 2). Most of those abnormalities had been previously identified by using the MLPA P335, P181, and P182 kits.
Figure 2.
Copy number variation (CNV) analysis by massively parallel sequencing. Log2 ratios of normalized mean coverage of each target were plotted against the reference. The x-axis is the corresponding targets in the panel, plotted by relative genome order, and the y-axis shows the log2 ratio of mean coverage of testing to that of reference. CNVs were called using fixed thresholds representing the minimum log2 ratio for gains (0.55) and maximum log2 ratio for losses (−0.55).
3.1.2. Archer® FusionPlex® ALL Kit
This is a panel intended to detect gene fusions using RNA as a template. In this study, the panel was sequenced on two different platforms, Illumina and Ion Torrent.
We confirmed that the total number of RNA reads per sample was not significantly different when the Archer® FusionPlex® ALL kit was run on two different platforms (Illumina or Ion Torrent) (p = 0.8).
The detection of the rearrangements was concordant between platforms in 4 out of 7 rearrangements detected. Sample 1 was known to harbor the t(9;22)(q34;q11), which was detected by the panel regardless of the platform. Sample 2 presented an LMO1-RIC3 fusion identified by both platforms. A BCL11B rearrangement was also detected when sequenced with an Illumina platform but was not found on Ion Torrent. On the other hand, an IKZF1 exon skipping was called by Ion Torrent but was not found when the panel was sequenced with Illumina equipment. No rearrangements were found in sample 3 harboring a dic(9;20)(p11-13;q11), although a PAX5 fusion was suspected. ETV6-RUNX1 rearrangement, previously confirmed by routine diagnostic techniques, was detected in sample 4, and a GGYF2-PBX1 fusion was also identified in this sample by both platforms. When this panel was sequenced on an Ion Torrent, a RUNX1-ETV6 fusion was also identified in sample 4.
Although the panel’s main purpose is to detect fusion genes, it is possible to detect SNVs when using an Illumina platform and analyzed using the Archer® analysis platform. In this case, we found a variant in SH2B3 in sample 2 and another variant in PTPN11 in sample 4. When the samples are sequenced on Ion Torrent and analyzed using IonReporter, the analysis of SNVs is not offered.
3.1.3. OncomineTM Childhood Cancer Research Assay (OCCRA)
Among the tested panels, this was the only panel able to detect SNVs and fusion genes by analyzing both DNA and RNA. Overall, OCCRA panel detected a mean of 1.75 SNVs per patient in 6 genes and 1 rearrangement in two patients.
This panel did not identify any SNV in sample 1; however, it detected a BCR-ABL1 fusion. Several variants were found in sample 2 in APC, SH2B3, TP53, and NF1, but no rearrangement was found. For sample 3, the panel identified a SNV in JAK2 and FLT3 and, as in sample 2, no rearrangement was found. Finally, a TP53 variant and ETV6-RUNX1 fusion were found in sample 4.
3.1.4. Human Comprehensive Cancer GeneRead Panel v2®
The Human Comprehensive Cancer GeneRead Panel v2® analyzes DNA to search for SNVs. This panel showed mutations in all the samples analyzed, with a mean of 2.75 variants per patient in 10 genes.
Specifically, this panel showed mutations in PIK3R1, PALB2, and ASXL1 genes in sample 1. Sample 2 harbored variants in MSH, APC, and TP53. In sample 3, the panel identified SNVs in JAK2, EP300, and TP53. Sample 4 presented mutations in PTPN11 and TP53.
3.2. Panel Dissection
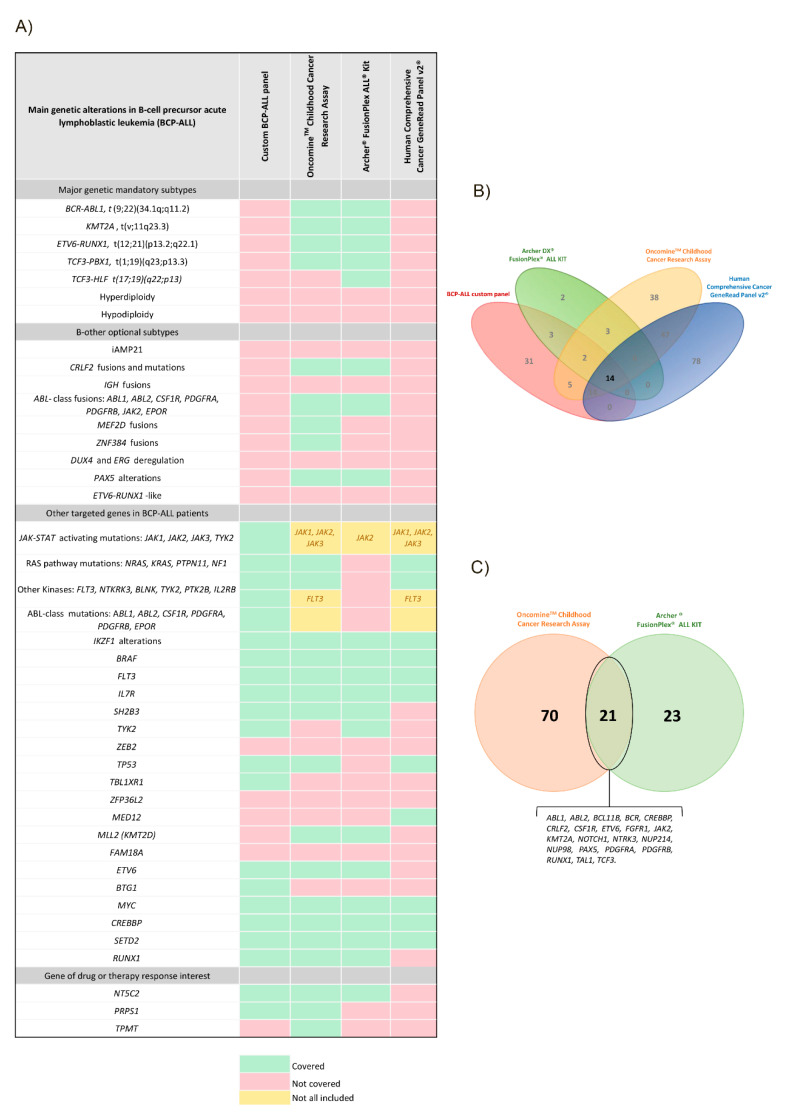
Main genetic alterations with clinical relevance for pediatric BCP-ALL diagnosis, overlapping gene content, and overlapping fusion content among panels are represented in Figure 3A–C, respectively.
Figure 3.
Gene content of the different targeted panels. (A) Approach of the different panels to assess the main genetic alterations in BCP-ALL. (B) Overlapping gene content in the different Next Generation Sequencing-targeted compared panels. (C) Overlapping fusion content in the OncomineTM Childhood Research Assay and Archer® FusionPlex® ALL kit.
All four panels had fourteen overlapping genes for SNV (BRAF, CREBBP, CRLF2, EZH2, FLT3, IL7R, JAK1, JAK2, JAK3, KRAS, NF1, NRAS, PAX5, and PTPN11). These 14 genes are known to be involved in ALL. The remaining genes present in the different panels were useful for different aims. Three of the four panels were specifically designed to improve childhood BCP-ALL diagnostics, as the genes included were relevant in pediatric cancer or ALL. The custom BCP-ALL panel was the most specific to search for mutations in pediatric ALL (Figure 3A) as it was designed for this project with this objective but did not cover gene fusions. Archer® FusionPlex® ALL kit was focused on fusion gene identification in ALL. OCCRA was designed to identify mutations and fusions in pediatric cancer, including solid tumors and hematological malignancies. Finally, the Human Comprehensive Cancer GeneRead Panel v2® had a broader approach, as it was conceived for the study of different types of cancer, mostly focused on adult cancer, but only detected mutations. Thus, two of the four panels, OCCRA and Archer® FusionPlex® ALL kit, were able to detect fusion genes. A total of 21 fusions were common among the panels. However, the design of all four panels did not include the study of highly relevant fusion genes in the BCP-ALL diagnostic: the custom BCP-ALL panel and Human Comprehensive Cancer GeneRead Panel v2® did not cover gene fusions, while Archer Fusion Plex comprised some of them (Figure 3A).
3.2.1. SNV Detection
Filtered Variant Call Format (VCF) obtained from the different in-house analysis for custom BCP-ALL panel or software analysis for commercial panels were compared. The variants called in each panel are shown in Supplementary Table S7. After variant calling, a similar number of variants per patient were called by the different panels. However, some differences were observed in the gene variant detection since the genes covered in each panel are different.
When focusing on the more common genes for SNV detection between the panels, variant calling and filtering was comparable. Correlation of the VAFs detected by each panel showed a high concordance level between panels for SNV detection.
Only the variant reporting process showed small discordances among centers for the custom BCP-ALL panel (described below). Thus, although similar variants and VAF were reported individually by different sequencing centers, some differences in reported variants were found (Figure 1 and Supplementary Table S6). In sample 1, the ETS2 variant found by centers 1 and 2 was not reported by center 3, as it was considered a polymorphism after finding a MAF > 1% in 8 different databases. In sample 2, center 2 missed the APC variant due to the low depth of coverage of this region (only 23 total base reads). In sample 3, three FAT3 variants were reported by center 2, but not by centers 1 and 3, as these variants were considered polymorphisms and filtered out during variant prioritization. Similarly, centers 1 and 3 reported a variant in FLT3 that was not reported by center 2, as it was considered a polymorphism, being reported in 2 databases with a MAF > 1% and presenting only one entry in the COSMIC database. The TP53 variant in sample 4 was only reported by center 2. In this case, discordance was attributed to differences in the transcript choice. The centers used two different transcripts, which had a major effect on the ultimate variant annotation. Thus, center 2 used transcript NM_001126117.1 to annotate this variant, obtaining an exonic missense variant in TP53 (p.(Ser213Leu); c.638C > T). In contrast, center 1 and 3 used transcript NM_000546.5, thus obtaining an intronic variant (c.993 + 309C > T), which was filtered out during the variant prioritization.
There were no differences between commercial panel reporting because the same software analysis was used.
3.2.2. CNV Detection
Since only the custom panel was designed to detect CNVs, no comparison between panels was performed.
3.2.3. RNA Fusion Detection
Our study showed that both assays that covered fusion detection, the Archer® FusionPlex® ALL kit and OCCRA, successfully detected fusions with known partners, previously identified by RT-PCR.
The total number of RNA reads did not differ significantly between the Archer® FusionPlex® ALL kit and the OCCRA panel (p = 0.3). The detection of rearrangements was concordant in 2 out of 4 samples. No rearrangements were found in sample 3, harboring a dic(9;20)(p11-13;q11). The ETV6-RUNX1 rearrangement, previously confirmed by routine diagnostic techniques, was detected in sample 4 by both commercial panels. On the other hand, sample 1 was known to harbor the t(9;22)(q34;q11)/BCR-ABL1, which was detected by both commercial panels, although the number of BCR-ABL1 transcript reads differed between platforms (a mean of 5067 reads on Archer® FusionPlex® ALL kit vs. a mean of 245,820 reads on OCCRA). The Archer® Fusionplex® ALL kit found the LMO1-RIC3 fusion in sample 2 in two independent runs (centers 2 and 4), whereas the OCCRA panel did not detect this transcript, as it is not included in its design.
All RNA results are shown in Supplementary Table S8.
4. Discussion
The use of NGS technologies has dramatically increased in clinical laboratories, as therapeutic protocols demand the screening of multiple genetic abnormalities with diagnostic, stratification, or therapeutic relevance. Besides, NGS has helped in the identification of targets for MRD follow-up. However, choosing the optimal NGS strategy for daily routine can be difficult. Genome-wide NGS approaches provide a high amount of information and are progressively cheaper, but the complexity in their analysis hampers their use in daily routine. NGS-targeted panels, in contrast, may represent a good option to implement in clinical practice. In our collaborative study, we assessed different currently available NGS-targeted approaches to apply in the setting of pediatric BCP-ALL diagnosis. We designed a custom panel and assessed its performance and that of three different NGS commercial panels. Overall, we demonstrated similar results for the different platforms and panels and confirmed their utility in a clinical context. However, some issues should be considered before implementing NGS in clinical practice in childhood ALL.
4.1. Panel Discussion
NGS panel target design depends on the panel’s intended use, as they can be designed with different scope. Collectively, the purpose and applicability of the different targeted panels tested were compared for their use in clinical laboratories (Supplementary Figure S2). The four panels had in common the presence of alterations described in BCP-ALL. The custom BCP-ALL panel had advantages in detecting SNV and CNV in selected genes related to pediatric BCP-ALL. OCCRA combined SNV and fusion detection and was focused on pediatric cancer-related genes. Archer® FusionPlex® ALL, based on its semi-anchored PCR, allowed to identify known fusion genes and discover new fusion partners, which may also be useful for research applications. On the other hand, the Human Comprehensive Cancer GeneRead v2® panel screened a large number of genes.
In our study, the four sequencing centers showed successful performance and concordance in results, not only in terms of variant detection but also in the VAFs obtained for each sample. However, different platforms showed some differences in fusion detection using the same commercial panel, probably due to the differences in the analytical software provided by the different companies. These results highlight the necessity of verifying the results by other methods. Our data confirm our custom panel’s reproducibility in different centers with the same platform, but not the full comparability of NGS platforms with different chemistries using the same panel. As used in this study, different NGS panels and platforms demonstrated comparable performance in the detection of somatic variants from DNA samples across multiple genes and a wide range of VAFs.
In terms of hands-on, all the NGS-targeted panels required time due to their extensive protocols consisting of various PCR steps. Our study tested one hybridization capture targeted panel (BCP-ALL custom panel) and 3 amplicon-based targeted panels (OCCRA, Archer® FusionPlex® ALL, and the Human Comprehensive Cancer GeneRead v2®). In general, amplicon-based panels are less time consuming than capture-based panels, shortening the time to get the results. Therefore, it is important to consider this point before choosing a panel to implement in daily routine. In terms of cost-effectiveness, generally, NGS panels are the most widely used tool for clinical applications mainly for cost-effectiveness reasons, as they allow a comprehensive study of the disease. However, different tests are still needed to identify all relevant genetic abnormalities contemplated in the therapeutic protocols.
Finally, it was not the scope of our study to demonstrate the clinical impact of the panels analyzed. Instead, we focused on each approach’s feasibility and performance and used a very limited number of cases. We selected four patients harboring already known clinically relevant abnormalities in terms of risk stratification and the prognostic impact such as TP53 and NF1 variants and BCR-ABL1 and ETV6-RUNX1 rearrangements. The commercial panels targeting RNA detected the rearrangements successfully, and all the DNA-based NGS panels correctly identified the variants, provided that adequate coverage of those regions was achieved (Supplementary Table S7).
Some factors may limit our study. A higher number of samples would be desirable. We also did not compare the different bioinformatics processes used by the different assays because of the significant differences in the panel design, library preparation, and sequencing platform. Thus, a truly fair comparison was not possible in our study, although it could be technically meaningful. Our study did not intend to carry out a technical validation of the panels; rather, we took the approach of a qualitative comparison to dissect the panels as a first step before choosing a panel. We also planned this study as the basis to harmonize the implementation of an NGS panel for the study of pediatric BCP-ALL in different centers in our country. However, a thorough validation of panels, including commercial assays, is needed before their implementation in clinical diagnostics.
4.2. Implementing NGS in Pediatric BCP-ALL
4.2.1. The Panel Choice
Before implementing NGS for clinical management of childhood leukemia, each center’s logistics, including available platforms, human, and economic resources, should be considered (Supplementary Figure S2). Additionally, comprehensive sequencing and integrative genome-wide analyses have profoundly refined the risk-stratification of pediatric BCP-ALL, resulting in the identification of new entities with prognostic and therapeutic significance [11]. Thus, the gene content may influence the panel choice and the clinical protocol should primarily guide this process.
Most commercially targeted NGS panels are designed as pan-cancer panels and contain a large number of genes. According to the literature, not all genes included in the panels have been shown to be clinically relevant, and, in the case that any clinically relevant gene was missed or not correctly covered, alternative methodologies should be performed to discard such abnormalities.
In this line, we have detected that any NGS panel is still facing at least two challenges in the BCP-ALL field: the detection of rearrangements, which is not fully covered in some currently available panels, and the detection of SNVs.
In this regard, the Human Comprehensive Cancer GeneRead v2® covers a vast range of cancer-related genes, so it could be useful for those laboratories that study a wide span of cancers; however, in a clinical laboratory focused on a specific type of cancer, as hematological neoplasms, could not be the best option. On the other hand, the custom BCP-ALL, the OCCRA, and the Archer® FusionPlex® ALL panels were designed focusing on pediatric cancer and ALL. The core of genes shared by all panels for SNVs (BRAF, CREBBP, CRLF2, EZH2, FLT3, IL7R, JAK1, JAK2, JAK3, KRAS, NF1, NRAS, PAX5, and PTPN11) covered mainly those genes involved in new classification subtypes (such as Ph-like or BCR-ABL1-like, and PAX5 alterations subtypes), which in turn represent new therapeutic targets involved in deregulated cell pathways in ALL [11,12,13,14]. Mutations in those genes related to BCP-ALL are pathogenically important and confer a better understanding of the disease. Thus, genetic testing might help clinicians tailoring personalized treatment to pediatric ALL patients. However, additional genes highly relevant to BCP-ALL were not included in the four panels design, such as TP53, CSF1R, or NF1. Therefore, when choosing an NGS panel for BCP-ALL, it might be important to prioritize the panel that includes the relevant genes with diagnostic, prognostic, and/or predictive value according to the current therapeutic protocols.
Interestingly, the main advantage of those panels that cover SNVs and fusion gene detection like the OCCRA panel (see Figure 3) is their ability to detect a wide percentage of abnormalities for accurate diagnostic classification, and the capacity to establish a precise patient risk stratification according to novel prognostic classification [13]. Of note, the Archer® FusionPlex® ALL open-ended PCR approach can detect previously unknown fusion genes and any partner of promiscuous genes; this is especially relevant in the BCP-ALL setting, as it allows to detect clinically relevant fusions involving KMT2A gene, as well as heterogeneous rearrangements involved in the Ph-like subtype.
Classical diagnostic techniques, such as karyotyping, PCR, and FISH, remain the gold standard in clinical routine in BCP-ALL diagnosis. Additionally, other conventional methods, such as MLPA and SNP-arrays, are often used to identify CNVs and larger chromosomal losses/gains. However, all the techniques mentioned above may miss identifying several alterations that have been recently reported by NGS approaches in the disease. Therefore, it is important to choose a panel covering a broad spectrum of BCP-ALL alterations, especially those new aberrations not detected by conventional methods.
4.2.2. Technical Challenges of Each Analyzed Panel for BCP-ALL Diagnosis
Several methodological issues may be found in each step of targeted NGS. First is the sample and library preparation. In this regard, the BCP-ALL custom panel is laborious and time-consuming, in contrast with the Archer® FusionPlex® ALL, the easiest hands-on protocol of all the panels tested. The second is the sequencing and data analysis for variant detection. The user has to take into account if the chosen panel has technical support, like Archer® FusionPlex®, or requires a specialist to develop a pipeline workflow. Another issue is the variant prioritization and reporting. Thus, we may find sequence errors due to artifacts that originate from library preparation, the sequencing process itself, or data analysis (e.g., read mapping, variant calling), resulting in misinterpretation of sequence variants [15]. Supplementary Table S9 summarizes the most significant challenges and possible approaches to tackle them.
Considering the hands-on in the optimization of our custom panel, we confirmed that the availability of commercial NGS panels allows an easier and faster application to the routine practice, as the panel performance has been previously tested and optimized by the company. However, even commercial panels need to be validated before their use in the clinical routine, as previously described.
Another issue to consider when choosing an NGS panel is the turnaround time expectation for the results, as it may play a crucial role in patient care. Considering the low incidence of pediatric BCP-ALL, the number of patients to analyze per week can be insufficient to perform a library each week. In this case, each laboratory should consider different options: (1) combining patients with different pathologies (BCP-ALL plus other types of leukemia, or solid tumor) using the same panel, as long as the scope of the panel allows it; (2) multiplexing different panels in the same run, or (3) using lower-capacity sequencing cartridges, studying only 1 or 2 patients per run. These approaches may allow shortening the turnaround time and providing the results in a clinically useful manner.
4.2.3. Data Analysis for Variant Detection
Differences in data analysis with bioinformatics pipelines and variant prioritization are often ignored as a source of discrepancies. In this line, we observed some discrepancies due to different variant annotation and prioritization criteria used among centers. It is also important to emphasize that the harmonization of NGS panels must be accompanied by standardized bioinformatics algorithms to optimize inter-center reporting reproducibility. Some authors propose using bioinformatics pipelines independent of those offered by the sequencing platform manufacturers (often heterogeneous and poorly adapted to specific requirements) [16].
In this context, the genome sequence used as a reference for mapping and transcript for variant annotation is another crucial step [17]. Differences in the annotation for one variant may lead to discrepancies in the clinical results. An example is a variant found in TP53 in sample 4 in our study. In this case, the use of different transcript sets resulted in the annotation for the same variant as intronic or exonic. This emphasizes the need for standardization of the annotation process to avoid the overstudy of potential disease-relevant variants and the omission of interesting variants incorrectly annotated. In the BCP-ALL setting, this issue is particularly relevant in some genes such as TP53, where mutations may occur throughout the gene [7].
4.2.4. Variant Prioritization and Reporting
To harmonize the NGS workflow between different centers, the criteria for prioritizing the detected variants should also be considered. The filtering of germline variants, the use of different databases and cut-offs for total reads, and VAF reporting need to be agreed upon. Initially, we let each center use their analysis pipelines and variant prioritization criteria (setting only minimum common criteria) so that we could evaluate if these possible discrepancies truly exist in the results. With our study, we can assert the need for standardization in crucial steps and reach a consensus on the pipelines, reference genome, and variant prioritization criteria.
Finally, variant interpretation and reporting in the clinical context of the BCP-ALL remain a difficult task. Detected variants should be carefully reviewed by appropriately trained and certified molecular diagnostic professionals and integrated into the context of other clinical and laboratory findings (cytomorphology, immunophenotyping, cytogenetics, and molecular data). The molecular professional should report each variant using the current evidence derived from different American College of Medical Genetics and genomics/Associations of Molecular Pathology (ACMG/AMP) variant curation guidelines [18,19].
5. Conclusions: Lessons Learned and the Future Ahead
Previous publications have shown that NGS is a useful tool in the clinical context in the molecular characterization of leukemia [20,21,22]. In the era of personalized medicine, NGS may be useful, but not limited, to find targetable lesions in kinase-driven BCP-leukemias. It is crucial to assess the maximum number of potentially relevant molecular markers, as they could be used to refine the risk stratification and, if needed, to include patients in ongoing clinical trials with new drugs in different phases of the disease. Besides, the molecular MRD monitoring will probably expand in the future as more markers are arising in a clonal manner [23,24,25].
The application of NGS technology in clinical diagnostics should proceed with care. As there is no consensus yet on a universal methodology for NGS, a constant interaction between molecular experts, technicians, and clinicians is essential for selecting the right NGS panel at diagnosis, the sequencing methodology, and the correct interpretation of the NGS results. Moreover, the analytical validation of the chosen panel is a prerequisite for using NGS technology in clinical routine, both individually and among different centers in cooperative groups [26]. Our study highlights the importance of a comprehensive dissection of available NGS panels before implementing them into the clinics.
The interaction between clinical and molecular experts in our collaborative study amongst Spanish centers helped to define the minimum set of useful genes in pediatric BCP-ALL, fostered the collaborative research, and provided a step forward toward a consensus on NGS technologies, which will lead to their optimized use for the benefit of patients.
Acknowledgments
We are indebted to our patients and families and to Obra Social from Hospital Sant Joan de Déu for their help. Samples and data from patients included in this study were processed following standard operation procedures with appropriate approval of the Ethical and Scientific Committees. We thank the Biobanc from Hospital Infantil Sant Joan de Déu per a la Investigació, Biobanco del Hospital Universitari i Politècnic La Fe (Biobanco La Fe, PT13/0010/0026), Basque Biobank (www.biobancovasco.org), and Biobanco from Hospital Universitario Virgen del Rocío, all integrated into the Spanish Biobank Network of Instituto de Salud Carlos III (ISCIII), for the samples and data procurement. Finally, we would like to thank the Group of Leukemia (not registered) of the Spanish Society of Pediatric Hematology and Oncology (SEHOP) for their support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4426/10/4/244/s1, Figure S1: Overview of the study: workflow of samples, panels, and sequencing platforms, Figure S2: Application of different targeted-gene panels for BCP-ALL, Table S1: Main characteristics of the BCP-ALL patients included in the study, Table S2: List of the 69 genes included in the BCP-ALL custom panel, Table S3: Content of Archer®FusionPlex® ALL Kit (ArcherDX, Inc., Boulder, CO, USA), Table S4: Content of OncomineTM Childhood Cancer Research Assay (ThermoFisher Scientific, Waltham, MA, USA), Table S5: List of genes of Human Comprehensive Cancer GeneRead Panel v2® (Qiagen, Hilden, Germany), Table S6: Single-nucleotide variants reported by each center in the common samples after variant prioritization, using the BCP-ALL custom panel, Table S7: Single-nucleotide variants reported by each center in each of the common samples after variant prioritization, using the BCP-ALL custom panel and three commercial panels, Table S8: Fusion genes identified in the common samples analyzed with the commercial panels Archer® FusionPlex® ALL and OCCRA on different platforms, Table S9: Critical steps in NGS procedures: challenges and possible approaches to face them. Sample and library preparation.
Author Contributions
N.V.-G., R.B., M.L., J.A., J.M.H.-R., M.R., and M.C. designed the study and M.C. supervised the project; N.V.-G., R.B., M.L., C.R., J.A., E.B., J.M.H.-S., A.M. (Alfredo Minguela), A.M. (Adrián Montaño), M.O., M.T., J.C., J.S., A.J.-V., and M.R. designed the customized panel; N.V.-G., E.E.-C., C.V.-G., M.L., M.M.-I., and C.R. performed the libraries and the sequencing process; G.F., J.M.H.-S., M.M.-I., and J.M. performed the bioinformatics analysis and N.V.-G., M.L., J.M.H.-S., and J.A. analyzed the data; N.V.-G., E.E.-C., C.V.-G., M.L., R.B., A.M. (Alfredo Minguela), A.M. (Adrián Montaño), and M.C. wrote the paper with the contribution of J.A., G.F., J.M., S.R. (Susana Riesco), and M.R., S.R. (Susana Riesco), J.M.H.-R., Á.L., J.M.F., S.R. (Susana Rives), and J.L.D. recruited patients. Other centers and investigators contributing to this study and belonging to the Biological Committee and other integrants of the Group of Leukemia of the Spanish Society of Pediatric Hematology and Oncology (SEHOP) are listed in the Supplementary Information. All authors have read and agreed to the published version of the manuscript.
Funding
This study has been supported by a grant from “Fundación Unoentrecienmil”; grants from Instituto de Salud Carlos III (PI12/02417; PI16/00246; PI12/00816; PI16CIII/00026; PI16/0665); Asociación Pablo Ugarte (TPY-M 1149/13; TRPV 205/18), ASION (TVP 141/17); Fundación Sonrisa de Alex & Todos somos Iván (TVP 1324/15); Consejería de Educación, Junta de Castilla y León (SA271P18); Fondos FEDER (CIBERONC [CB16/12/00284]), Proyectos de Investigación del SACYL, GRS 1847/A/18, Generalitat Valenciana GV2019/077 and Fundación AMPILE. J.M.H.S. is supported with a research grant by FEHH (“Fundación Española de Hematología y Hemoterapia”), A.M. a grant from Junta provincial de Salamanca de la Asociación Española Contra el Cáncer (AECC), and M.M. holds the grant “Ayuda predoctoral de la Junta de Castilla y León” by the Fondo Social Europeo (JCYL-EDU/556/2019 Ph.D. scholarship).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hunger S.P., Mullighan C.G. Acute Lymphoblastic Leukemia in Children. N. Engl. J. Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 2.Pui C.-H., Yang J.J., Hunger S.P., Pieters R., Schrappe M., Biondi A., Vora A., Baruchel A., Silverman L.B., Schmiegelow K., et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J. Clin. Oncol. 2015;33:2938–2948. doi: 10.1200/JCO.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greaves M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2018 doi: 10.1038/s41568-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pui C.-H., Gaynon P.S., Boyett J.M., Chessells J.M., Baruchel A., Kamps W., Silverman L.B., Biondi A., Harms D.O., Vilmer E., et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet. 2002;359:1909–1915. doi: 10.1016/S0140-6736(02)08782-2. [DOI] [PubMed] [Google Scholar]
- 5.Gatta G., Rossi S., Foschi R., Trama A., Marcos-Gragera R., Pastore G., Peris-Bonet R., Stiller C., Capocaccia R. Survival and cure trends for European children, adolescents and young adults diagnosed with acute lymphoblastic leukemia from 1982 to 2002. Haematologica. 2013;98:744–752. doi: 10.3324/haematol.2012.071597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haslam K., Catherwood M.A., Dobbin E., Sproul A., Langabeer S.E., Mills K.I. Inter-Laboratory Evaluation of a Next-Generation Sequencing Panel for Acute Myeloid Leukemia. Mol. Diagn. Ther. 2016;20:457–461. doi: 10.1007/s40291-016-0222-3. [DOI] [PubMed] [Google Scholar]
- 7.Burghel G.J., Hurst C.D., Watson C.M., Chambers P.A., Dickinson H., Roberts P., Knowles M.A. Towards a Next-Generation Sequencing Diagnostic Service for Tumour Genotyping: A Comparison of Panels and Platforms. BioMed Res. Int. 2015;2015:478017. doi: 10.1155/2015/478017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathology M., Area H., Service P., Dna N., Meldrum C., Doyle M.A., Tothill R.W. Next-Generation Sequencing for Cancer Diagnostics: A Practical Perspective. Clin. Biochem. Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 9.Bastida J.M., Lozano M.L., Benito R., Janusz K., Palma-Barqueros V., Del Rey M., Hernández-Sánchez J.M., Riesco S., Bermejo N., González-García H., et al. Introducing high-throughput sequencing into mainstream genetic diagnosis practice in inherited platelet disorders. Haematologica. 2018;103:148–162. doi: 10.3324/haematol.2017.171132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knoppers B.M., Nguyen M.T., Sénécal K., Tassé A.M., Zawati M.H. Next-generation sequencing and the return of results. Cold Spring Harb. Perspect. Med. 2016;6:a026724. doi: 10.1101/cshperspect.a026724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inaba H., Mullighan C.G. Pediatric Acute Lymphoblastic Leukemia. Haematologica. 2020;105:2524–2539. doi: 10.3324/haematol.2020.247031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu Z., Churchman M.L., Roberts K.G., Moore I., Zhou X., Nakitandwe J., Hagiwara K., Pelletier S., Gingras S., Berns H., et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019;51:296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwab C., Harrison C.J. Advances in B-cell Precursor Acute Lymphoblastic Leukemia Genomics. HemaSphere. 2018;2:e53. doi: 10.1097/HS9.0000000000000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilljebjörn H., Fioretos T. New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood. 2017;130:1395–1401. doi: 10.1182/blood-2017-05-742643. [DOI] [PubMed] [Google Scholar]
- 15.Bacher U., Shumilov E., Flach J., Porret N., Joncourt R., Wiedemann G., Fiedler M., Novak U., Amstutz U., Pabst T. Challenges in the introduction of next-generation sequencing (NGS) for diagnostics of myeloid malignancies into clinical routine use. Blood Cancer J. 2018;8:113. doi: 10.1038/s41408-018-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sujobert P., Le Bris Y., de Leval L., Gros A., Merlio J.P., Pastoret C., Huet S., Sarkozy C., Davi F., Callanan M., et al. The Need for a Consensus Next-generation Sequencing Panel for Mature Lymphoid Malignancies. HemaSphere. 2019;3:e169. doi: 10.1097/HS9.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy D.J., Humburg P., Kanapin A., Rivas M.A., Gaulton K., Cazier J.-B., Donnelly P. Choice of transcripts and software has a large effect on variant annotation. Genome Med. 2014;6:26. doi: 10.1186/gm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Leng W.W.J., Gadellaa-Van Hooijdonk C.G., Barendregt-Smouter F.A.S., Koudijs M.J., Nijman I., Hinrichs J.W.J., Cuppen E., Van Lieshout S., Loberg R.D., De Jonge M., et al. Targeted next generation sequencing as a reliable diagnostic assay for the detection of somatic mutations in tumours using minimal DNA amounts from formalin fixed paraffin embedded material. PLoS ONE. 2016;11:e0149405. doi: 10.1371/journal.pone.0149405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-García G., Baux D., Faugère V., Moclyn M., Koenig M., Claustres M., Roux A.F. Assessment of the latest NGS enrichment capture methods in clinical context. Sci. Rep. 2016;6:20948. doi: 10.1038/srep20948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alonso C.M., Llop M., Sargas C., Pedrola L., Panadero J., Hervás D., Cervera J., Such E., Ibáñez M., Ayala R., et al. Clinical Utility of a Next-Generation Sequencing Panel for Acute Myeloid Leukemia Diagnostics. J. Mol. Diagn. 2019;21:228–240. doi: 10.1016/j.jmoldx.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Press R.D., Eickelberg G., Froman A., Yang F., Stentz A., Flatley E.M., Fan G., Lim J.Y., Meyers G., Maziarz R.T., et al. NGS-Defined Minimal Residual Disease Before Stem Cell Transplantation Predicts Acute Myeloid Leukemia Relapse. Am. J. Hematol. 2019;94:902–912. doi: 10.1002/ajh.25514. [DOI] [PubMed] [Google Scholar]
- 24.Kotrova M., Trka J., Kneba M., Brüggemann M. Is Next-Generation Sequencing the way to go for Residual Disease Monitoring in Acute Lymphoblastic Leukemia? Mol. Diagn. Ther. 2017;21:481–492. doi: 10.1007/s40291-017-0277-9. [DOI] [PubMed] [Google Scholar]
- 25.Qin X., Zhang M.-Y., Liu W.-J. Application of minimal residual disease monitoring in pediatric patients with acute lymphoblastic leukemia. Eur. Rev. Med. Pharmacol. Sci. 2018;22:6885–6895. doi: 10.26355/eurrev_201810_16158. [DOI] [PubMed] [Google Scholar]
- 26.Jennings L.J., Arcila M.E., Corless C., Kamel-Reid S., Lubin I.M., Pfeifer J., Temple-Smolkin R.L., Voelkerding K.V., Nikiforova M.N. Guidelines for Validation of Next-Generation Sequencing–Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.