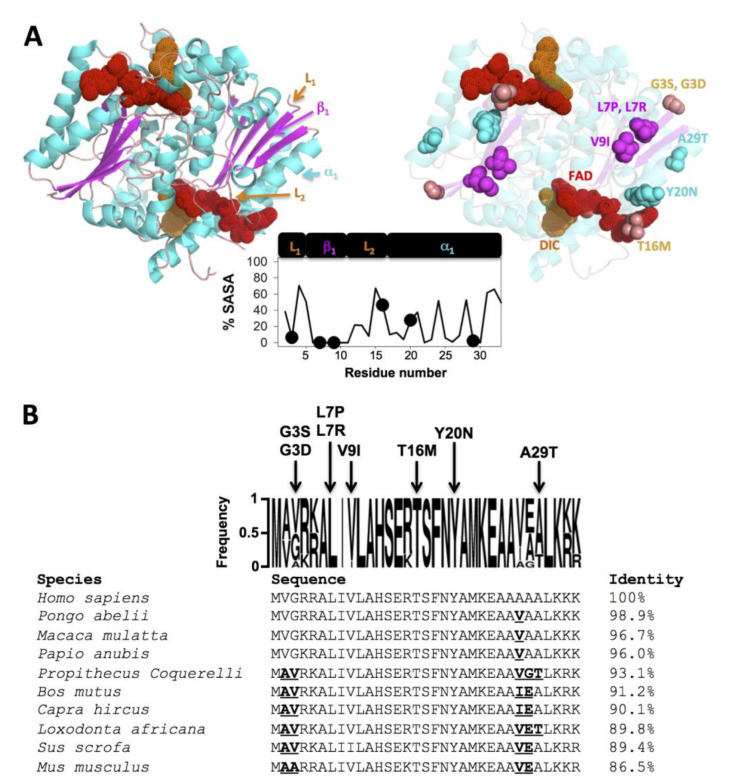
Figure 1.
Structural location and sequence conservation of the NQO1 mutations studied in this work. (A) Structural representation on the dimeric structure of NQO1 (PDB code 2F1O; [72]). The left panel shows the secondary structure elements to which mutated residues belong (L1, residues 1–4; β1, residues 5–10; L2, residues 11–16 and α1, residues 17–33). The right panel shows the location of the mutated residues as well as the FAD and dicoumarol (Dic) molecules. Residues are colored according to the secondary structure. The plot in the middle shows the solvent accessibility (% SASA) of this region (determined using GETAREA on the PDB code 2F1O as the average of the two monomers in the dimer). (B) Sequence alignment of 10 selected NQO1 mammalian proteins. Residues in bold-underlined indicate non-conservative mutations vs. the human sequence (note that K-to-R or V-to-I are considered as conservative). The frequency plot over the NQO1 alignment (generated using WebLogo, https://weblogo.berkeley.edu/) also shows the identity of the missense mutations investigated in this work.