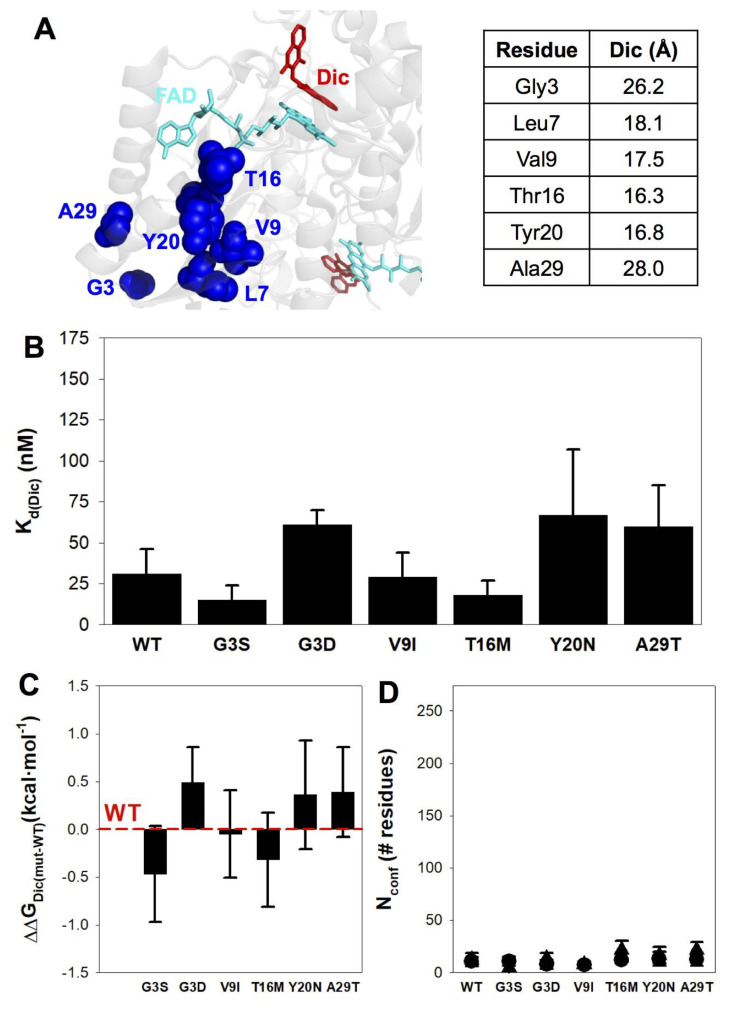
Figure 6.
Affinity and structure–energetics analysis for Dic binding to NQO1 variants. (A) Location of mutated residues and minimal distances to Dic (using PDB code 2F1O). (B) Dissociation binding constants at 25 °C. Data are mean ± s.d. from at least three independent experiments for each variant. (C) Difference in binding free energy between a given mutant and the WT protein. Errors are those determined from linear propagation. (D) Magnitude of the conformational change (as a number of residues, Nconf) determined from experimental binding enthalpies (circles) and changes in heat capacity (triangles).