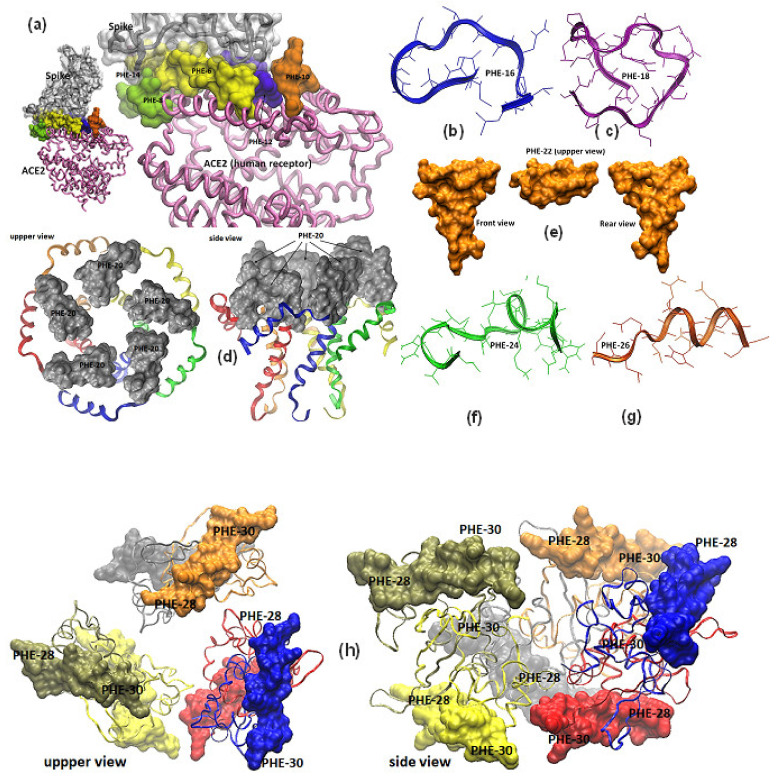
Figure 2.
Molecular models of SARS-CoV-2 surface proteins and relative location of designed epitope-peptide targets. (a) Representation of the SARS-CoV-2 spike receptor-binding domain complexed with angiotensin converting enzyme 2 (ACE2), protein data bank (PDB) file code 6m0j [7], epitope-peptides PHE-2, PHE-4, PHE-6, PHE,8, PHE-10, PHE,12 and PHE14 are highlighted in the interface of the 3D structure. PepFold molecular prediction models for epitope-peptides PHE-16 and PHE-18 from ORF3a are shown in blue and purple ribbons in (b,c) Pentamer envelope small membrane protein-E (PDB code 5 × 29) of a formed ion channel resolved by NMR experiments, in which the epitope-peptide PHE-20 is displayed as a gray surface in all protein chains [9]. (d) The M protein (membrane or matrix protein) is a triple-spanning membrane arranged, predictive M-based epitope-peptidePHE-22 3D structure was analyzed by PepFold and represented as a yellow mustard solid surface. (e) Modified epitope-peptides PHE-22 and PHE-24 based on the expression product of ORF7a were molecular modelized by the PepFold predictive 3D structure algorithms and herein represented as green and orange ribbons in (f,g). (h) Dimers associated with a hexamer complex 3D structure (PDB codes 6m3m and 6wkp) were resolved by X-ray crystallography experiments for the nucleocapsid RNA binding domain of N protein and herein is represented as colorful ribbons in which peptides PHE-28 and PHE-30 are highlighted as solid surfaces.