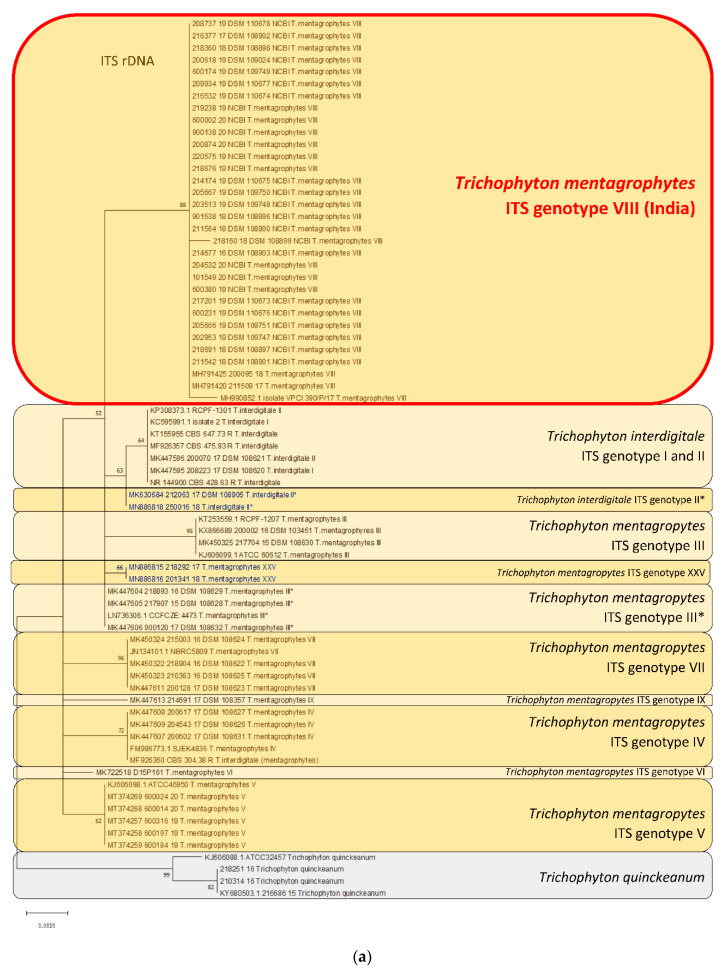
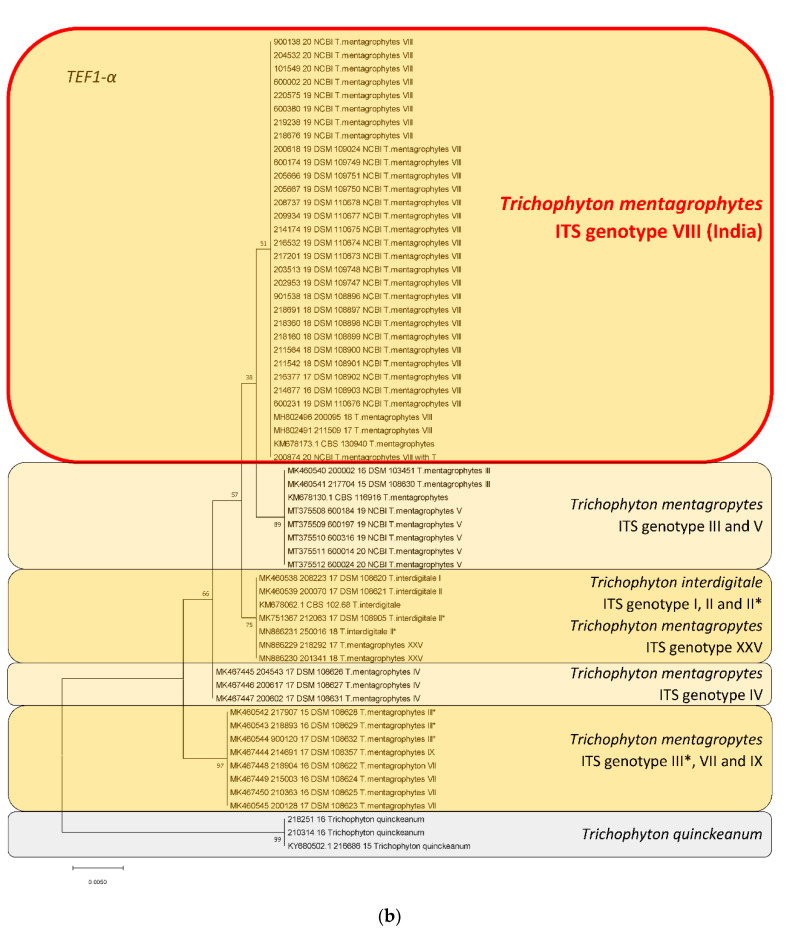
Figure 4.
Phylogenetic analysis of the dermatophyte isolates based on the ITS regions of rDNA and the TEF1-α gene (Table 2).The evolutionary history was inferred by using the maximum likelihood method and Tamura-Nei model [10]. The tree with the highest log likelihood (−907.70) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 28 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. There were a total of 1086 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [11]. (a) Phylogenetic tree of T. mentagrophytes based on sequencing of the ITS regions of rDNA genes. By sequencing, a 100% concordance with NCBI reference strains (accession numbers MH791420, MH791425, MH990852) was found for all 29 isolates. All these isolates formed their own cluster, which is now called the ITS genotype VIII (India) of T. mentagrophytes. These isolates (T. mentagrophytes Type VIII or clade) were clearly discriminated from already known T. mentagrophytes genotypes, e.g., II, V, VII. Rooted with Trichophyton quinckeanum. (b) Phylogenetic tree of T. mentagrophytes based on sequencing of the TEF1-α gene. Used NCBI reference sequences (TEF1-α gene) were MH802491 and MH802496 (accession numbers). Within the phylogenetic tree, all T. mentagrophytes ITS Type VIII strains from Germany formed their own clade, which is clearly discriminated from the other, above mentioned, T. mentagrophytes genotypes. Rooted with T. quinckeanum.