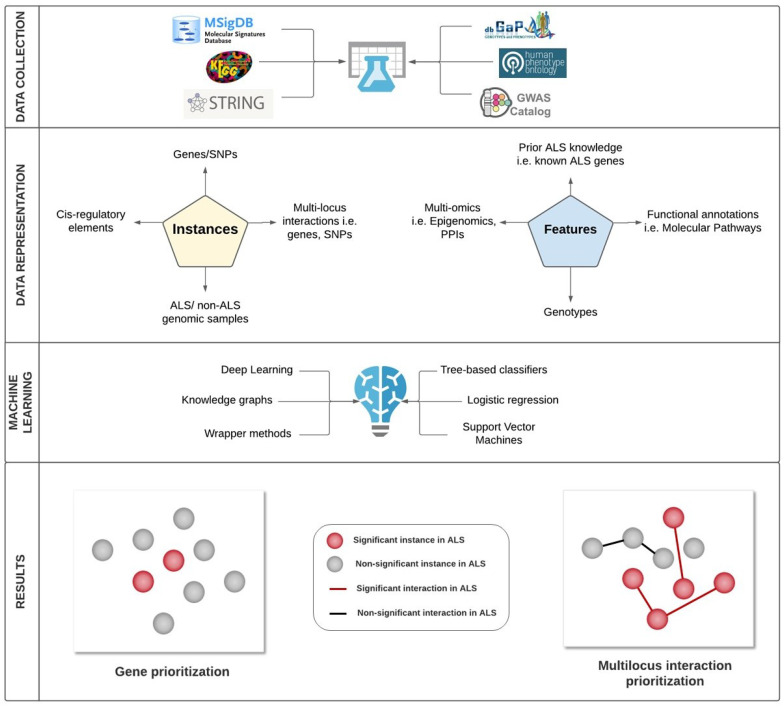
Figure 2.
Some of the key decision making steps taken by machine learning approaches in ALS genomics, and examples of some possible choices at each step. These included data collection, data representation, selection of the Machine Learning algorithm for the classification task, and the types of result obtained by the model. The studies collected information from a variety of databases in order to mine, among other things, data on genotypes (e.g., dbGaP, GWAS Catalog, gnomAD), functional annotations (e.g., KEGG), and Protein-Protein Interactions (e.g., STRING). Depending on the purpose of the experimental design, the collected studies modelled genes, SNPs, cis-regulatory regions, multilocus interactions, and/or ALS/non-ALS patients. Each instance was described using features such as Genomic, Epigenomic, and Proteomic data, functional annotations, and/or prior ALS-related knowledge (e.g., ALS gene-sets). Various machine learning algorithms were selected for the classification tasks. Lastly, we visualize the distinction between the modelling results of gene prioritization and multilocus interaction prioritization studies. Gene prioritization studies aim to identify significant ALS associated instances (e.g., SNPs, genes and cis-regulatory regions), whereas multilocus interaction prioritization studies aim to discover significant interactions among multiple loci. Lastly, we note that an ALS versus non-ALS sample classification experiment can be used to prioritize genes if the interpretability of the chosen model permits the identification of informative genomic features.