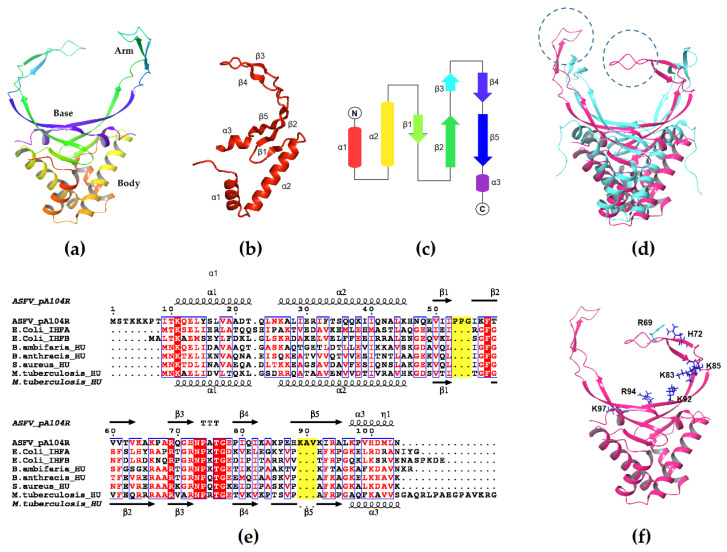
Figure 1.
The structure of pA104R: (a) Model of dimeric pA104R generated by the Schrödinger Maestro suite [59]; (b) One protomer of the dimeric pA104R; (c) Topological diagram of secondary structural elements of the pA104R protomer; (d) Model of pA104R (magenta) superimposed with M. tuberculosis HU (cyan). The lengthened beta 2–3 and 4–5 loops are encircled; (e) Multiple sequence alignment of pA104R with other HU/IHF members generated by Clustal Omega (version 1.2.4) [60] using the HHalign algorithm and its default settings as described in [61] and ESPript 3.x [62]. The extra two sets of amino acids are highlighted in yellow; (f) The positively charged amino acid residues in the bottom (K92, R94, and K97) and arm regions (R69, H72, K83, and K85) of the pA104R protomer that contribute the majority of the protein-DNA interactions. The PDB codes of the structures are 6LMH (pA104R) and 4PT4 (mtbHU). Accession numbers are P68742, P0A6X7, P0A6Y1, B1YQ53, A0A2A7DF13, Q99U17, and P9WMK7.