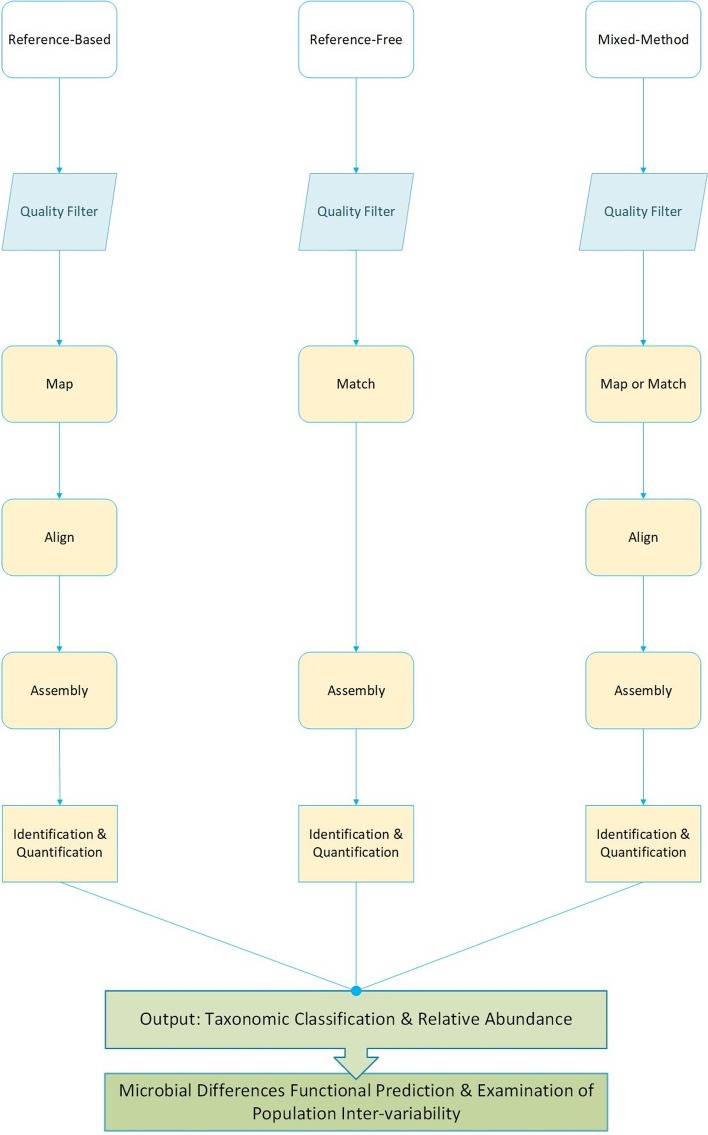
Fig. 1.
Generic pipeline comparing three basic computational frameworks designed to identify microbial reads from human sequences. Generic pipelines can be summarized into three general stages, pre-processing (blue), processing (yellow), and analyses post-processing (green). During pre-process, most methodologies trim and quality filter sequencing reads. Quality reads are mapped and aligned during the processing steps to either human or pathogen reference sequences or key identifying factors before making a final identification call. Once species have been identified, their composition is characterized in detail, depending on the methodology being used. Finally, having taxonomic classification and compositional structure permits downstream correlation analyses and functional-relevant identification of molecular pathways. Differential functional prediction and patient inter-variability aid in the identification of novel microbe based prevention and treatment strategies