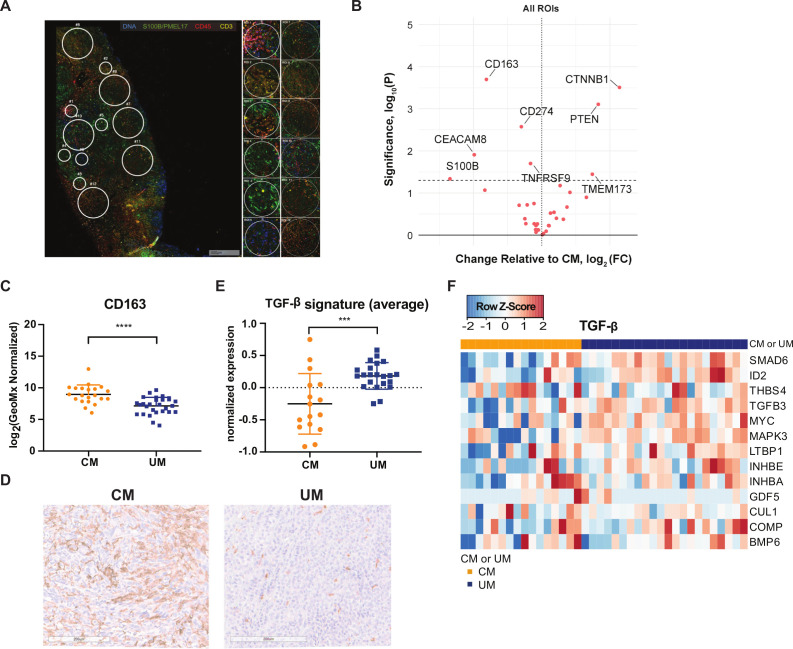
Figure 5.
Digital spatial profiling (DSP) analysis of cutaneous melanoma (CM) and uveal melanoma (UM) liver metastases. (A) Example of regions of interest (ROI) selection using the visualization makers syto13 (blue), S100B/PMEL17 (green), CD45 (red) and CD3 (yellow). Per patient 12 ROIs were selected in tumor-infiltrating lymphocyte (TIL) high, low or random areas. areas of 200 µm in diameter (n=6) and 600 µm in diameter (n=6) were placed. (B) Volcano plot of differently expressed markers by DSP analysis. Left: markers higher expressed in CM; right: markers higher expressed in µm. Dotted line p value cut-off. (C) DSP analysis of CM and UM liver metastases for CD163. The mean and SD are shown. (D) Representative immunohistochemistry for CD163 staining. (E) Average expression of transforming growth factor beta (TGF-β) gene signature for CM (orange) and UM (blue) patients. The mean and SD are shown. (F) Heatmap of transcription TGF-β gene signature.65 Columns represent patients (CM patients (orange); UM patients (blue)). Positive values (red) are indicated as higher expression, negative values (blue) are indicated as lower expression. ***p≤0.001, ****p≤0.0001.