Figure 3.
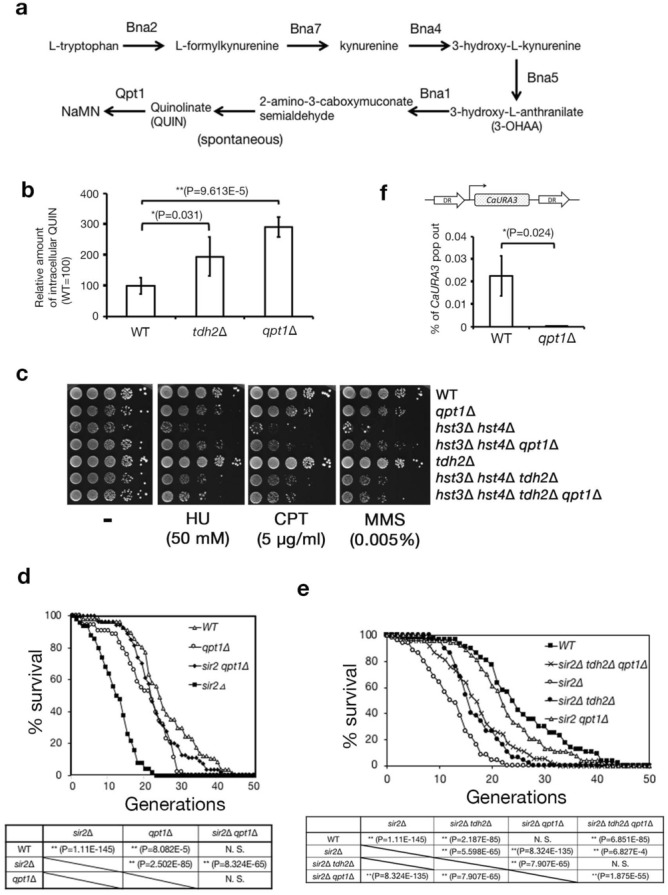
Quinolinic acid (QUIN) is a candidate metabolite that is increased in tdh2∆ cells to suppress replication fork slippage. (a) The metabolic pathway of the de novo NAD+ synthesis pathway (kynurenine pathway). NaMN: nicotinic acid mononucleotide. (b) Comparison of intracellular QUIN levels among strains. Relative amount of QUIN in each cell per wild-type cell (WT = 100). *P < 0.05. **P < 0.01. Unpaired t-test (two-tailed). Error bars represent the standard deviation of three biological replicates. (c) Plate assays monitoring the sensitivity of each DNA damaging agent. (d,e) Pedigree analysis to count replicative lifespans among cells. *P < 0.05. **P < 0.01. N.S: nonsignificant. Unpaired t-test (two-tails). Over 50 cells/strain were used for analysis. (f) The frequencies of the CaURA3 deletion among repeats. DRs: direct repeats. *P < 0.05. Unpaired t-test (two-tailed). Error bars represent the standard deviation of three biological replicates.
