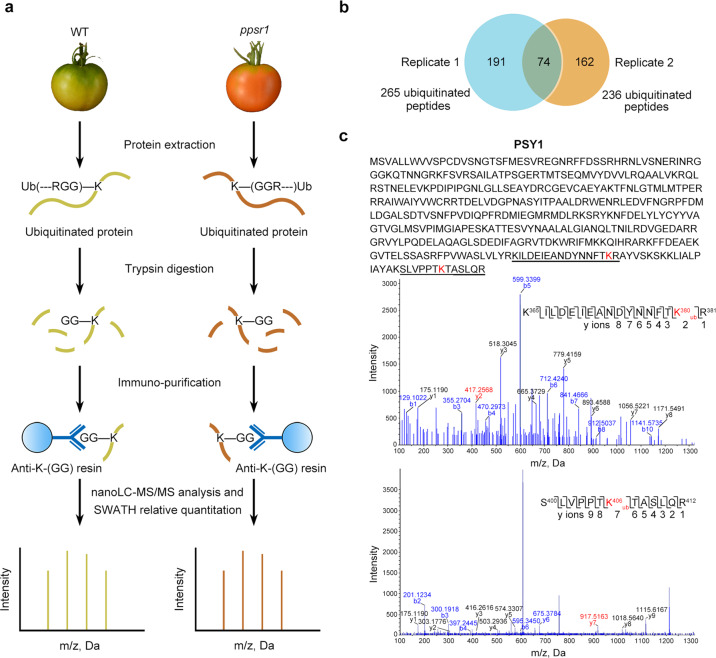
Fig. 4. Identification of PSY1 as the candidate substrate of PPSR1.
a A workflow diagram showing the identification and quantification of ubiquitinated peptides that exhibit significant differences in abundance in the ppsr1 mutant fruit compared to the wild type (WT). Proteins isolated from WT and ppsr1 mutant fruit at 38 DPA were digested with trypsin, followed by immunoprecipitation with an anti-K-(GG) antibody. The recovered ubiquitinated peptides were submitted to SWATH-MS (Sequential Window Acquisition of all Theoretical Mass Spectra) quantitative proteomic analysis. b Overlap of the ubiquitinated peptides identified in two independent biological replicates. c Identification of ubiquitination sites in PSY1 by NanoLC–MS/MS. Sequences of the identified ubiquitinated peptides are underlined in the PSY1 protein sequence. The mass spectra of two ubiquitinated peptides are displayed. The y-ions and the corresponding peptide sequence are presented, with ubiquitinated lysine (K) residue marked in red.