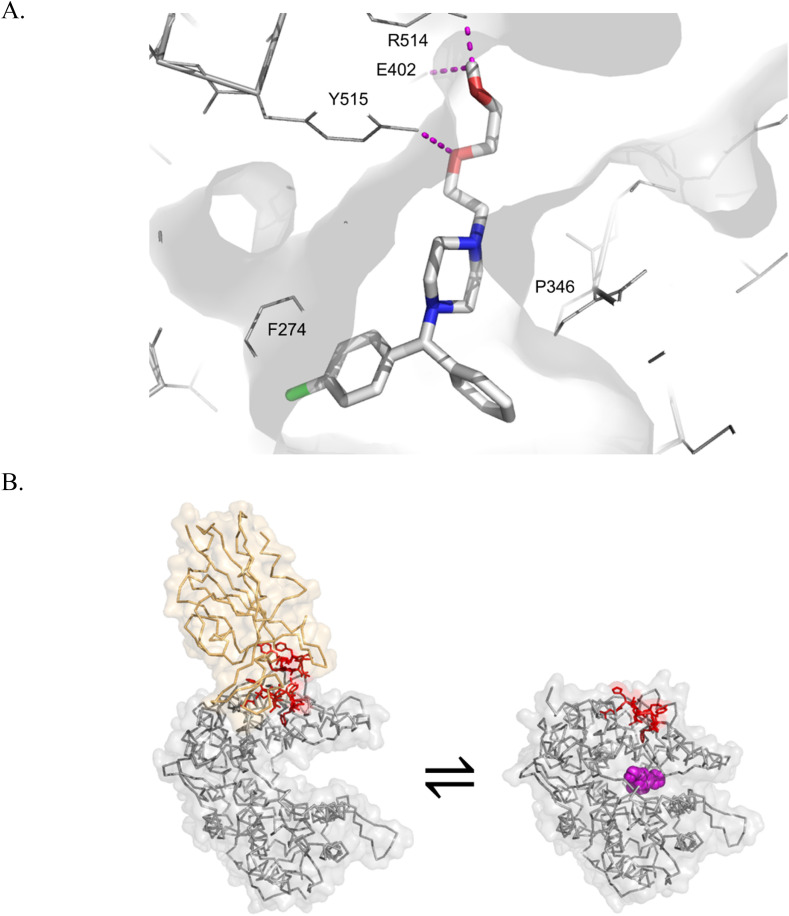
Fig. 2.
In silico modeling of the ACE2 catalytic inhibitor hydroxyzine suggests binding within the active site. A. Hydroxyzine was posed by molecular docking with AutoDock Vina. Hydroxyzine is shown as sticks, white for carbon, blue for nitrogen, red for oxygen, green for chlorine. Polar interactions (e.g., H bonds) shown as magenta dashes. B. The SARS-CoV-2 spike glycoprotein binds the open conformation of ACE2. The SARS-CoV-2 spike protein is shown in orange (from PDB 6M17). ACE2 is shown in grey in the open conformation (left, from PDB 6M17) and the closed conformation (right, PDB 1R4L). Inhibitor binding prevents intermolecular contacts between ACE2 and SARS-CoV-2 spike protein (shown as sticks in red). Hydroxyzine is shown as spheres in magenta, as posed by AutoDock Vina.