Figure 2.
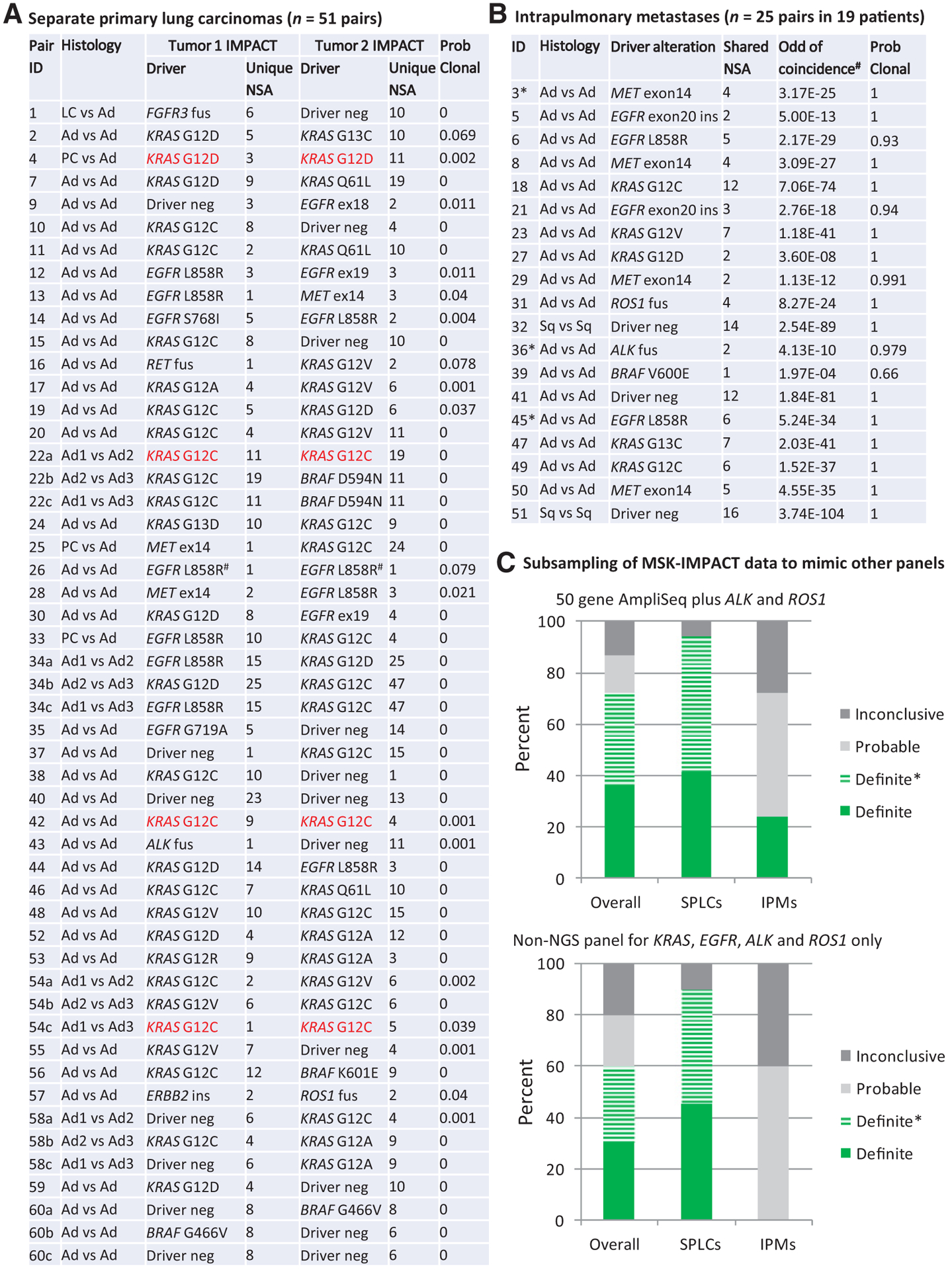
Distribution of driver alterations and subsampling to mimic more limited platforms. A, Driver alterations in 51 SPLC tumor pairs from 41 patients. Highlighted in red are SPLCs with coincidentally shared hotspot driver mutations that could be misclassified as favoring IPMs by other platforms. #, shared EGFR L858R in case 26 represents tumor pairs with different nucleotide changes in EGFR gene resulting in the same amino acid substitution. B, Driver alterations in 25 IPM tumor pairs from 19 patients.*, three IPM cases with asterisks (3, 36, and 45) denote patients with three tumors. C, Subsampling of MSK-IMPACT data to mimic performances of noncomprehensive molecular platforms. Definite SPLC tumor pairs showing different driver alterations in each tumor. Definite SPLC*, one tumor shows a driver alteration while the other lacks that alteration (See Supplementary Materials and Method 2). Definite IPM, ≥2 shared somatic alterations. Probable IPM, one shared somatic alteration. Inconclusive, neither tumor shows a driver alteration. Ad, adenocarcinoma; amp, amplification; fus, fusion; ins, insertion; LC, large-cell neuroendocrine carcinoma; NSA, nonsynonymous alterations; PC, pleomorphic carcinoma; Sq, squamous cell carcinoma.
