(
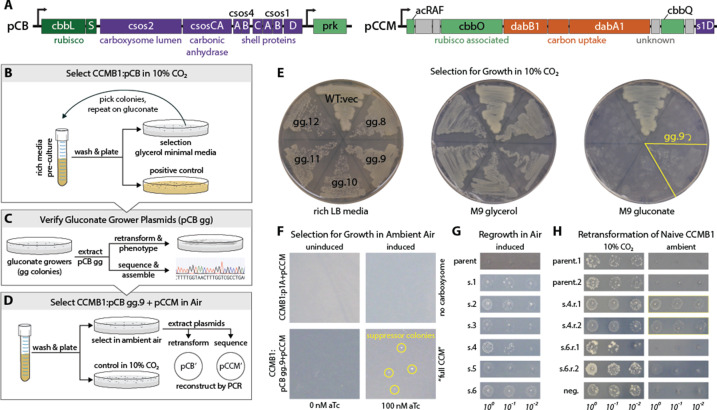
A) pCB and pCCM plasmids together encode 20
H. neapolitanus genes including 12 confirmed CCM components. pCB carries kanamycin resistance and has two transcriptional units both expressed under an aTc-inducible P
LtetO-1 promoter (
Lutz and Bujard, 1997). The first derives from pHnCB10 (
Bonacci et al., 2012) and expresses 10 carboxysome proteins. The second expresses phosphoribulokinase (
prk). pCCM carries chloramphenicol resistance and expresses an 11 gene operon from
H. neapolitanus that contains both putative and confirmed CCM genes (
Desmarais et al., 2019). Although pCB expresses both rubisco and
prk, CCMB1:pCB did not initially grow in M9 media under 10% CO
2 (not shown) and so we undertook a series of selections, described in panels (
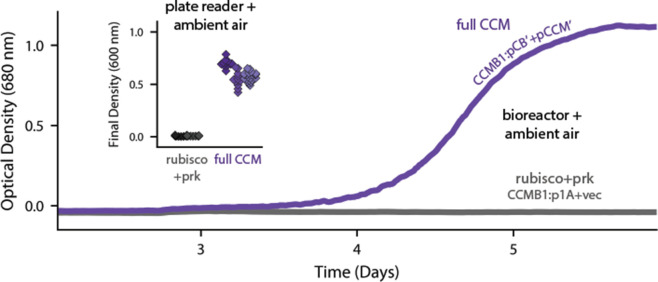
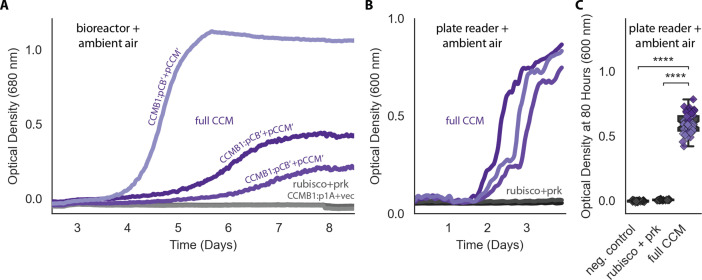
B–D) that ultimately led to isolation of pCB’ and pCCM’ plasmids that together enable CCMB1 to grow in ambient air. (
B) We first selected CCMB1:pCB for growth on minimal media by screening for mutants able to grow on M9 glycerol and then M9 gluconate media. Gluconate growing mutant #9 (gg.9) was used for subsequent experiments as this mutant was found to grow best on gluconate (as shown in E). (
C) Plasmid extracted from gg.9 was deep sequenced and electroporated into naive CCMB1 to test for plasmid linkage of growth on minimal. (
D) Selection for rubisco-dependent growth in ambient air. A turbid pre-culture of the CCMB1:pCB gg.9+pCCM double transformant was washed and plated on M9 glycerol media under ambient air. Colonies formed after ≈20 days (as shown in F). Forty colonies (s.1–40) were picked into rich media, grown to saturation, washed and plated on M9 glycerol media to verify growth under ambient air. Roughly ¼ of chosen colonies regrew under ambient air to varying degrees (s.1–6 are shown in G). Plasmid extracted from several strains was deep-sequenced and electroporated into naive CCMB1 to test plasmid-linkage of growth on glycerol minimal media in ambient air. Pooled plasmid extracted from s.4 was found to confer replicable growth in ambient air (as shown in H). PCR and Gibson cloning were used to reconstruct the individual pCB and pCCM plasmids from this pool. We termed these reconstructed plasmids pCB’ and pCCM’. (
E) Restreaking of gluconate-growing mutants gg.8–12 described in panel B shows that gg.9 grew best on gluconate. (
F) CCMB1:pCB gg.9+pCCM double transformants were plated for mutants on M9 glycerol media under ambient air. A negative control lacking carboxysome genes (CCMB1:p1A+pCCM) was plated at the same time. Colonies formed after 20 days (bottom right) only on induced plates (100 nM) and only when all CCM genes were provided (i.e. pCB gg.9 and pCCM). (
G) Several of the chosen colonies regrew in ambient air. Growth characteristics varied from colony to colony, suggesting genetic variation. (
H) Pooled plasmid extracted from s.4 was found to permit naive CCMB1 to grow in ambient air. For comparison, plasmid from s.6 produced less reproducible growth in ambient air.