Figure 1.
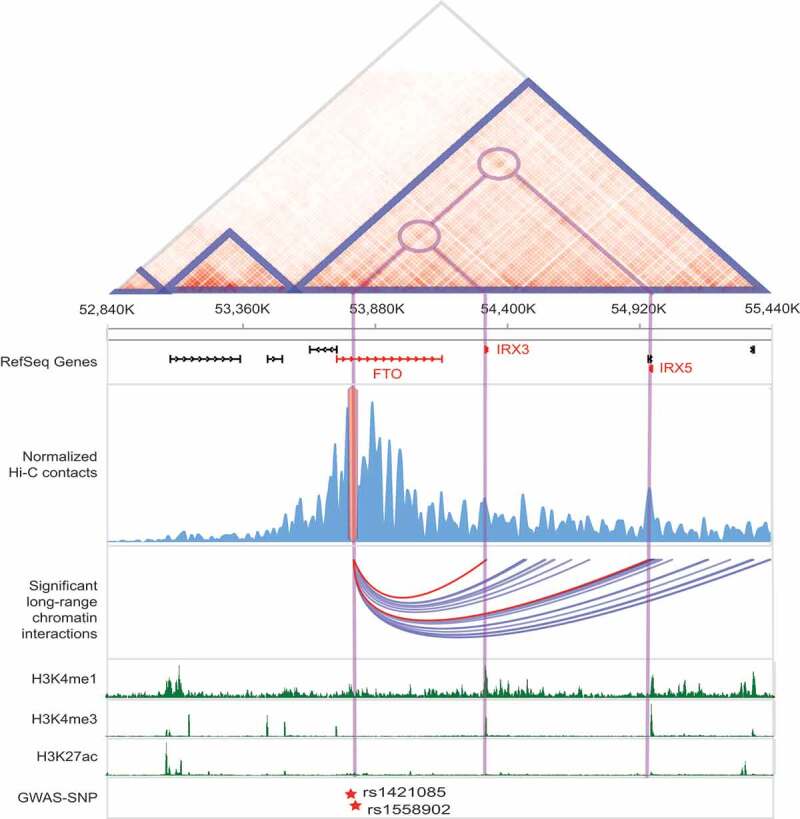
Target gene identification of cREs harboring obesity-associated GWAS-SNPs
Visualization of significant long-range chromatin interactions (arcs) centered on obesity-associated GWAS-SNPs (rs1421085 and rs1558902) located at FTO intron in H1-derived mesenchymal stem cell. From the top, normalized Hi-C contact map with annotation of TADs (blue triangles), RefSeq gene annotation, normalized Hi-C contacts with obesity-associated GWAS-SNPs located in intronic regions of FTO gene, identified significant chromatin interactions, multiple histone modification marks, and GWAS-SNPs are shown together. The long-range chromatin interactions with the promoters of IRX3 and IRX5 are highlighted by red. All data were drawn from 3DIV database [36].
