Figure 1.
Application of NucleusJ 2.0 autocrop method on a wide-field stack
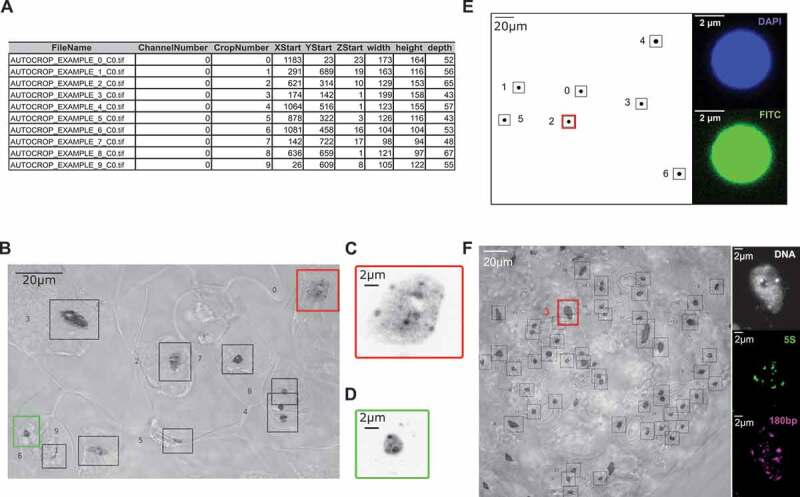
(a) Output table from a typical experiment ‘Autocrop_example’ using Arabidopsis cotyledons stained with Hoechst 33,258 DNA dye.(b) Overlay of a maximum Z-projection of the Autocrop_example wide-field stack gained from a sample of cotyledon stained with Hoechst 33,258 and a single plane image under transmission light using Differential Interference Contrast (DIC). Coordinates (X, Y and Z start) of the nine nuclei are described in Figure 1a. DIC allows identifying nuclei of guard cells (autocrop boxes #0 and # 6, respectively, bordered in red and green), which are located at stomates and pavement cells outside of stomates. Autocrop automatically draws bounding boxes (from 0 to n) of the overlay and inverted Lookup Table (LUT) to easily look at the nuclei position. This example also highlights rare cases where the bounding box contains two different nuclei (autocrop box #4).(c) and d) are close-up images of sub-regions of the Z-projection illustrating, respectively, a large pavement cell nucleus (autocrop box #0 in red) and a smaller and rounder guard cell nucleus (autocrop box #6 in green). (e) Multi-channel autocrop on 4 µm microspheres. Maximum Z-projection of a wide-field stack (left) with seven microspheres (#0 to 6). Selected Z-slice (right) of the same microsphere (autocrop box#2 in red) in two fluorescent channels; DAPI channel (top), FITC channel (bottom).(f) Multi-channel autocrop on plant nuclei. Overlay of a maximum Z-projection of the autocrop in DAPI channel wide-field stack (left) with 46 nuclei. Selected Z-slice (right) of nucleus #3 (red box) in three fluorescent channels; DAPI channel (gray level), Cy5 for 5S rDNA probe (green level) and Cy3 for 180bp probe (magenta level).
