Figure 3.
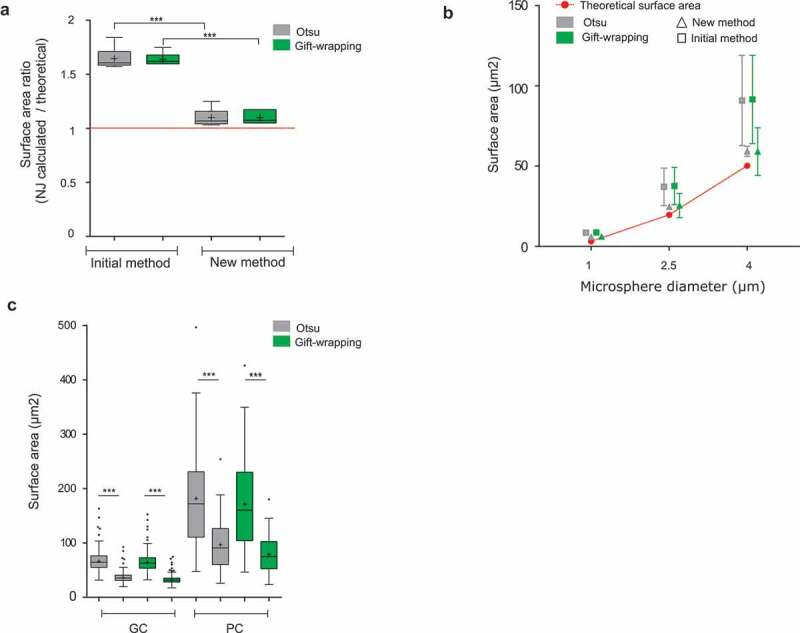
Evaluation of a new method of Surface Area calculation implemented into NucleusJ 2.0
(a) Digitized spheres of various radii of 5, 10, 20, 30, 40, and 50 voxels were generated and used to calculate the surface area (Supplemental table 1) with the initial and newly developed calculation method. Data are presented as a ratio between the observed and theoretical size of the digitized spheres. Student t-test P-value: *** <0.0001.(b) Fluorescent Microspheres of diameter of 1, 2.5, and 4 µm (n = 28, 24, and 15, respectively) were imaged using the wide-field microscope with an optigrid module, subjected to autocrop and segmented by the two NucleusJ 2.0 segmentation methods. Surface area gained from Otsu and gift-wrapping segmentation was then computed with the initial and new method of calculation (Supplemental table2). Red: theoretical surface area; gray: Otsu method; green: gift-wrapping method; triangle: new method; square: initial method. (c) Plant nuclei. Surface area of nuclei from cotyledon epidermis of WT plants were segmented by the Otsu or gift-wrapping methods. Nuclei from guard cells (GC, n = 375) and pavement cells (PC, n = 127) (Supplemental table 4). Student t-test P-value: *** <0.0001.
