Figure 1.
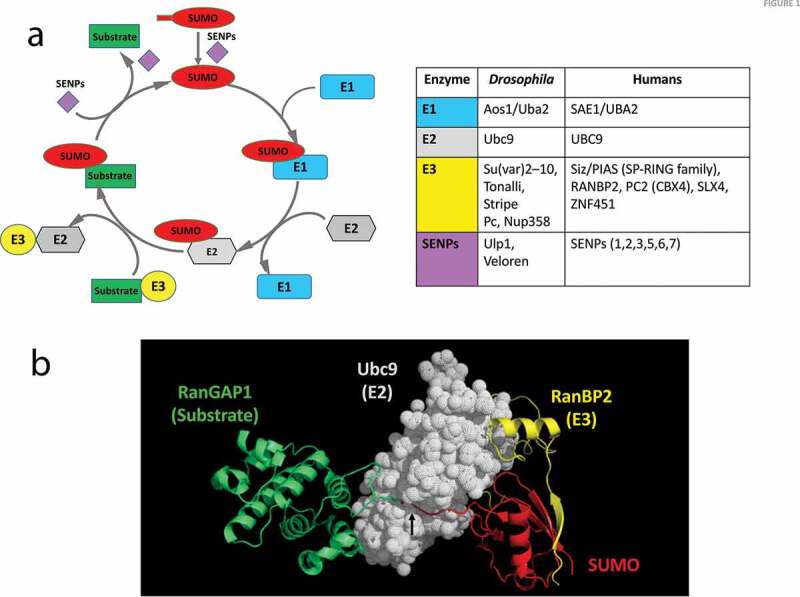
SUMO Conjugation of a substrate protein
(a) The SUMO conjugation/de-conjugation cycle. The addition and removal of SUMO to a target substrate is under enzymatic control. The first step is the maturation of SUMO by an endoprotease, named sentrin-specific protease (SENP) or Ubiquitin-like specific protease (ULP) that exposes the C-terminal di-glycine motif. Next, the E1 heterodimer engages with SUMO via a thioester linkage and subsequently hands it over to the E2 enzyme. The E2 then interacts with the substrate and catalyses the conjugation of the C-terminal COOH of SUMO to a specific lysine side-chain of the substrate. This conjugation step may be either enhanced or directed by an E3 ligase enzyme. The SENPs also serve to de-conjugate SUMO from the target, releasing it for a new cycle. The table lists the enzymes involved in regulating the SUMO cycle. Drosophila enzymes include putative E3 ligases inferred from homology with mammals and also gene ontology analysis. (b) Crystal structure (Protein data bank ID 1Z5S[48]) of SUMO conjugated to RanGAP1 (substrate) by the E2 (Ubc9), with RanBP2 acting as an E3 ligase. The structure shows the interaction between the substrate (RanGAP1) and Ubc9, as well as the cleft/tunnel (black arrow) in Ubc9 that holds the C-terminal GG tail of SUMO for conjugation with the lysine side chain of RanGAP1. The figure was generated using coordinates from the PDB using PyMol (The PyMOLMolecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
