Figure 3.
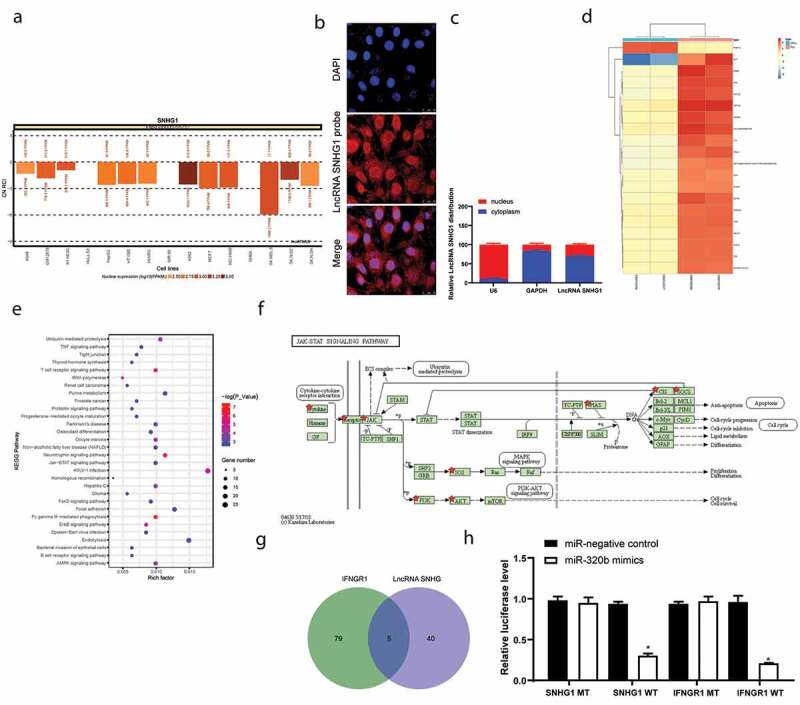
SNHG1 serves as a ceRNA for miR-320b to mediate IFNGR1 expression. A, subcellular localization of SNHG1 predicted on the lncATLAS DataBase (http://lncatlas.crg.eu/); B, FISH experiments with probes targeting SNHG1 were performed to validate the subcellular localization of SNHG1 in LFCs. The cytoplasm was stained with probes targeting SNHG1 (red stain), and the nuclei were stained with DAPI (blue stain); C, nuclear and cytoplasmic expression of SNHG1 in LFCs determined by QRT-PCR; D, heatmap for the top 30 differentially expressed genes between PLL- and OPLL- LFCs analyzed using the Llimma Rstudio based on GSE5464 microarray, with |LogFC ≥ 4.0| and p < 0.05 as the screening criteria; E, KEGG-based clustering analysis was performed on DAVID 6.8 software, and a total of 43 pathways where the differentially genes clustered were figured out; F, genes clustered on the JAK/STAT signal pathway; G, intersected miRNAs that could bind to SNHG1 and IFNGR1 predicted on StarBase; H, binding relationships between SNHG1 and miR-320b and between miR-320b and IFNGR1 validated by dual luciferase reporter gene assay. The data are expressed as the mean ± SD; two-way ANOVA and Tukey’s multiple comparison test was used to determine statistical significance; *, p < 0.05
