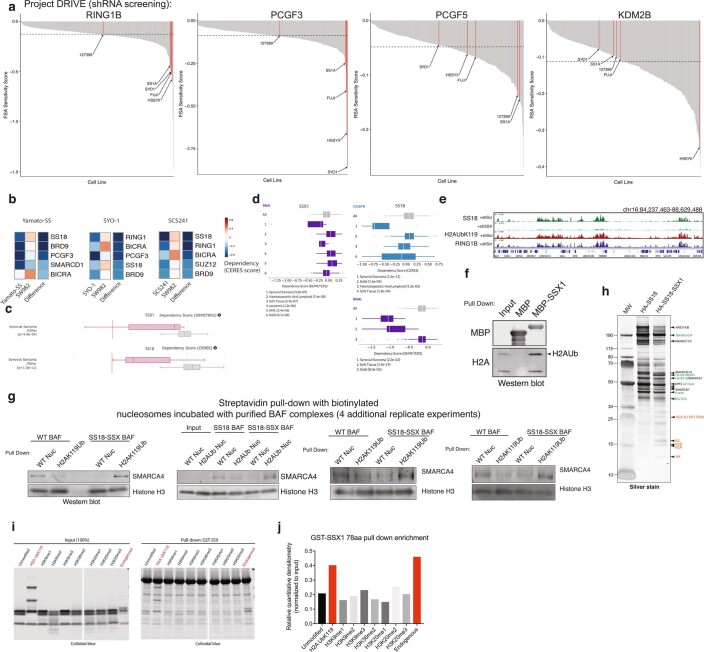
Extended Data Fig. 8. SS18-SSX-bound BAF complexes preferentially bind H2AK119Ub-marked nucleosomes.
a, Waterfall dependency plots for RING1B, PCGF3, PCGF5 and KDM2B genes across n = 387 cell lines in Project DRIVE Dataset (Novartis). b, CERES dependency scores (fitness dropout) derived from genome-scale fitness screens performed using CRISPR-Cas9-based methods (Achilles, Broad Institute; https://depmap.org/portal/achilles/). Difference is the score calculated between SYO1, Yamato-SS, SCS241 (SS18-SSX + ) cells and SW982 cells (negative for fusion, histologic mimic). Blue, enriched for dependency. mSWI/SNF, PRC1, PRC2 members are shown. c, CERES and DEMETER Dependency scores for SSX1 and SS18 genes for CRISPR-Cas9 and RNAi datasets, respectively. Synovial sarcoma cell lines are indicated in pink; all other cell lines are represented in gray. d, CERES and DEMETER Dependency scores for SSX1 and SS18 genes for CRISPR-Cas9 and RNAi datasets, respectively. Synovial sarcoma and soft tissue (SS cell lines) exhibit preferential dependency. (Project DRIVE; https://oncologynibr.shinyapps.io/drive/). SS cell lines containing the SS18-SSX fusion oncoprotein are highlighted in red. e, H2AK119Ub and RING1B ChIP-seq tracks over selected loci, aligned with SS18 (BAF) localization in SYO1 cells treated with shScramble or shSS18-SSX. f, MBP-SSX1 (78aa) pull-down experiments indicate capture of histones, and specifically, H2AK119Ub species. g, Streptavidin-based pull-down experiments (n = 4 independent replicate experiments) using endogenous, fully-assembled HA-SS18- or HA-SS18-SSX-bound BAF complexes incubated with biotinylated WT nucleosomes (unmodified) or H2AK119Ub-modified nucleosomes. SMARCA4 and Histone H3 immunoblots are shown. h, Representative silver stain of the WT SS18 complexes and SS18-SSX fusion complexes isolated using ammonium sulfate nuclear extraction protocol. Identified proteins are labeled. i, Pull-down experiments performed using GST-SSX incubated with unmodified or a series of modified recombinant mononucleosomes, or endogenous mononucleosomes (mammalian, purified via MNase digestion from HEK-239T cells). j, Quantitative densitometry performed on experiment in Extended Data Fig. 8i. Uncropped gel images for all relevant panels are presented in Supplementary Figure 1.