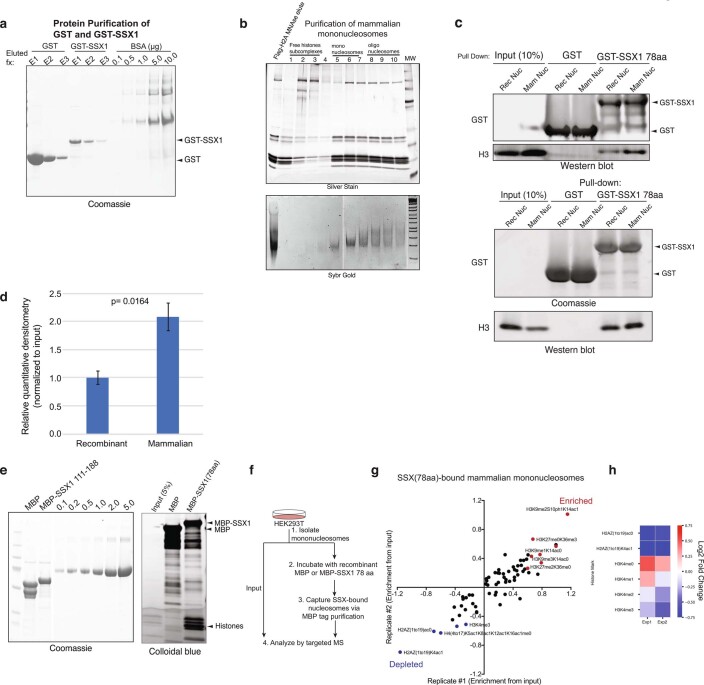
Extended Data Fig. 2. The SSX 78aa protein binds mononucleosomes, with preference for nucleosomes decorated with repressive histone modifications.
a, Coomassie-stained gel of recombinantly-purified GST, GST-SSX (78aa) proteins, run next to BSA protein as control. b, Purification of mammalian mononucleosomes from HEK293T cells using MNase digestion. c, Incubation of GST or GST-SSX (78aa) with either recombinant or mammalian mononucleosomes, resolved by immunoblot for GST and histone H3 or Coomassie and histone H3. Two representative experiments are shown. d, Quantitative densitometry of histone H3 normalized to input reflecting GST-SSX 78aa preferential binding to mammalian mononucleosomes (prepared via MNase digestion in HEK293T cells) versus recombinant, unmodified nucleosomes. Bars represent averages of n = 3 independent experiments, error bars represent mean ± s.d.; p-value= 0.0164 using two-tailed Student’s t-test. e, Purification of MBP and MBP-SSX (78aa) proteins for targeted, quantitative histone mass-spectrometry. f, Schematic for targeted MS experiments. g, Enrichment of SSX-bound histone peptides, over input. Enriched and depleted proteins are shown in red and blue, respectively. Quantitative histone mass spectrometry performed on MBP-SSX1 (versus MBP control) incubated with pooled mononucleosomes isolated from HEK293T cells via MNase digestion. Experiment performed in n = 2 replicates. See also Supplementary Table 2. h, Heatmap reflecting enrichment or depletion of selected histone marks, including H2AZ and H3K4 methylation states. Scale= log2FC (normalized to input), blue (negatively enriched), red (positively enriched). See also Supplementary Table 2. Uncropped gel images for all relevant panels are presented in Supplementary Figure 1. Data for panels 2d, 2g-2h are available as source data.