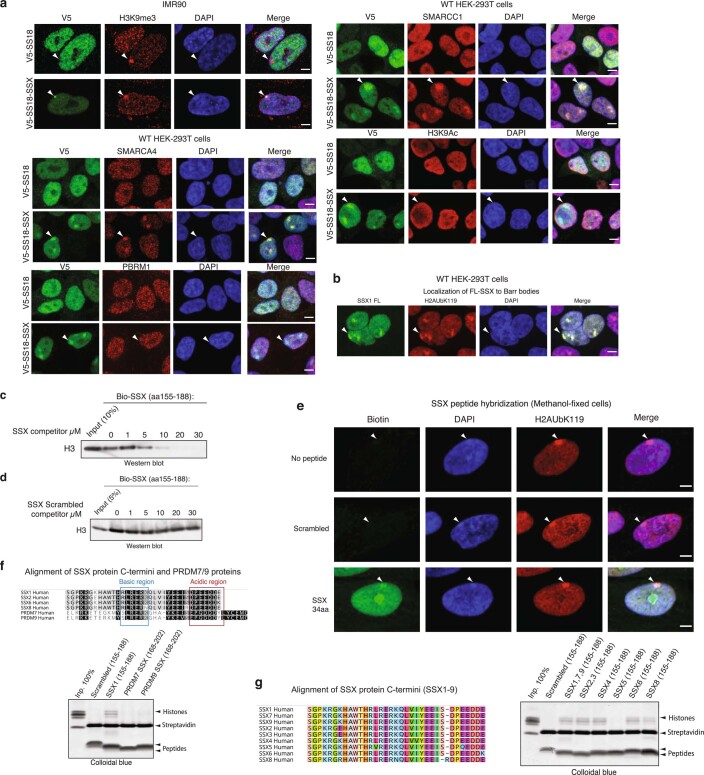
Extended Data Fig. 3. Nucleosome binding and nuclear localization properties of SS18-SSX and SSX variants.
a, Immunofluorescence imaging performed on IMR90 fibroblasts and HEK293T cells infected with either V5-SS18-SSX or V5-SS18. Visualized in red for H3K9me3, SMARCA4, PBRM1, SMARCC1, H3K9Ac across experiments. DAPI is shown as nuclear stain and merged images are provided with scale bars, Scale bar, 5μm. b, IF-based localization of SSX1 FL (1-188aa) in fibroblasts. H2AK119Ub, DAPI counterstain, and merged images are shown. Scale bar, 5μm. c, Peptide competition experiment using Biotinylated SSX peptide (aa 155-188) and unlabeled SSX (aa 155-188). Visualization for Histone H3 using immunoblot. d, Peptide competition experiment using Biotinylated SSX peptide (aa 155-188) and Scrambled control SSX peptide (aa 155-188). Visualization for Histone H3 using immunoblot. e, SSX peptide hybridization experiments performed on methanol-fixed cells. Streptavidin (SA) used for visualization of biotinylated SSX peptides, H2AK119Ub for Barr bodies. DAPI counterstain and merged images shown. Scale bar, 5μm. f, Top, conservation analysis among SSX and PRDM7 and PRDM9 human protein regions. Bottom, peptide pull-down experiments with recombinant nucleosomes performed with Scrambled control SSX1, SSX1, PRDM7, PRDM9. Visualization by colloidal blue staining. g, Left, alignment of SSX proteins (SSX1-9). Right, peptide pull-down experiments with recombinant nucleosomes performed with aa 155-188 of SSX family members. Visualization by colloidal blue staining. Uncropped gel images are presented in Supplementary Figure 1.