Abstract
The AT-rich Interactive Domain 1A (ARID1A) is one of the most frequently mutated genes in gastric cancer. Here, we found that genetic variants in noncoding regions of ARID1A associated with altered protein levels by target sequencing. Notably, tumors with ARID1A variants in the 3’untranslated region (3’UTR) exhibited remarkably increased heterogeneity of ARID1A protein. In general, genetic variants and protein deficiency of ARID1A in tumors were associated with a better survival. Strikingly, altered patterns and heterogeneity of ARID1A protein expression were observed in peritumor tissues and carried significant implications in defining tumor immune contexture by multiplex immunohistochemistry. By analyzing the spatial distribution of TILs, we showed that reduced ARID1A protein levels in both tumor and peritumor tissues were significantly correlated with increased density and proximity of TILs to tumor cells. In contrast, high heterogeneity of ARID1A expression was associated with increased TIL density, but reduced proximity of TILs to tumor cells. Collectively, our study characterized ARID1A genetic alterations and its protein expression patterns in EOGC, demonstrating new strategies for clinically assessing its molecular impact on tumor onset and progression, tumor immune response, and patient survival.
Keywords: ARID1A, target sequencing, early-onset gastric cancer, heterogeneity, tumor immune contexture
Introduction
ARID1A (AT-rich interaction domain 1A, also called BAF250) is a key component of the ATP-dependent chromatin remodeling complex SNF/SWI. It has an evolutionarily conserved function in regulating cellular processes associated with chromatin compaction such as gene transcription, DNA replication and DNA repair [1-4]. Genomic sequencing data has identified ARID1A as one of the most frequently mutated genes across diverse human cancers [5,6]. In TCGA dataset (cBioPortal for Cancer Genomics) [7], high mutation rates of ARID1A were found in endometrium-related carcinomas including more than 50% ovarian clear cell carcinoma (OCCC), more than 30% of ovarian endometrioid carcinoma and in about 40% of uterine endometrial carcinoma [8,9]. ARID1A mutations are also present in more than 30% of gastric carcinoma and urothelial bladder carcinoma [5,10,11]. In addition, 10-15% colorectal carcinoma, hepatocellular carcinoma and cholangiocarcinoma contain ARID1A mutations [12-15]. The majority of identified ARID1A mutations were inactivating nonsense or frame shift mutations, which resulted in loss of ARID1A expression [8,9]. Decreased and absent ARID1A protein expression was further confirmed in human tumor samples by immunohistochemistry analysis [9]. Studies in a variety of animal models have further demonstrated that ARID1A is a bona fide tumor suppressor through regulating gene transcription, genome maintenance mechanisms and cell proliferation/differentiation. Given the high frequency of ARID1A mutations in tumors, studies have identified and developed potential therapeutic strategies to target ARID1A deficiency including inhibitors of histone deacetylases and inhibitors of DNA damage response kinase ATR and DNA repair enzyme poly [ADP-ribose] polymerase (PARP) [16-18]. More recently, ARID1A loss has been associated with the alterations in tumor infiltrating lymphocytes (TILs) and treatment responses to immune checkpoint blockade through regulating mismatch repair-mediated mutation load and transcription-mediated interferon (IFN) signaling [6,19,20]. These studies suggested that ARID1A mutation/deficiency in tumors might be a potential biomarker for stratifying patients for targeted and immune therapy.
Notwithstanding findings of ARID1A as a key tumor suppressor and potential therapeutic target, important gaps in knowledge remain concerning ARID1A mutations and deficiency in tumors. First, ARID1A gene is located in the genomic region of 1p36.11. The ARID1A DNA sequence contains 86080 bp and coding mRNA contains 8595 bp. It contains 20 exons coding a protein product of 2285 amino acids. The current mutation spectrum of ARID1A aberrations was primarily localized in the coding region due to the sequencing technology specifically targeting exomes or lack of coverage depth in whole-genome sequencing [7,10,11,21-24]. A previous study reported that 5% of OCCC lacked protein expression without ARID1A coding mutations suggesting the potential that additional as yet uncharacterized mutations affect ARID1A [9]. Thus, it remains to be determined whether deleterious ARID1A mutations may occur in non-coding regions.
Second, although inactivating mutations such as nonsense or frameshift mutations were frequently found in ARID1A coding regions, a considerable number of mutations were identified in tumors that retained detectable protein expression [25]. The effect of specific alterations in ARID1A gene on its protein expression particularly missense mutations remains to be further examined.
Third, current studies analyzing the correlation between ARID1A protein levels and molecular changes in tumor immune microenvironment such as TILs were primarily focused on ARID1A expression in tumor tissues. In addition to cancerous tissues, ARID1A mutations and protein deficiency have been found in premalignant lesions such as endometrial hyperplasia with atypia and also in benign inflammatory lesions of endometriosis, which are strongly associated with endometrium-related carcinomas [26-29]. These data indicated that ARID1A loss could be an early molecular event during tumorigenesis. Histologically normal peritumor tissues are integral components of tumor microenvironment. It remains largely unexplored whether peritumor tissues contain ARID1A loss and whether altered ARID1A expression in peritumor tissues may impact on shaping immune responses in tumors.
Fourth, consistent with these findings of ARID1A deficiency in non-cancerous lesions with high risks of developing cancer, tissue-specific knockout mouse models in multiple cancer types including ovarian, breast, liver and pancreatic cancers demonstrated that ARID1A deficiency promotes the initiation of tumorigenesis through cooperating activation of oncogenic signaling (PI3K and K-Ras mutations) and loss of tumor suppressors (p53 and PTEN) [30-35]. These studies indicated that ARID1A loss not only facilitates tumor progression, but also drives tumor onset. However, whether ARID1A mutations and deficiency are associated with cancer diagnosed in young patients (early-onset cancers) remains unknown. It has been postulated that due to less environmental carcinogen exposure, early-onset human cancers provide an ideal background to study genetic changes at the initiating stages of tumorigenesis.
ARID1A mutations were found in more than 30% of GCs in the TCGA. There are marked genetic, proteomic and clinicopathological differences between early-onset GCs (EOGCs) (under 40 years old) and traditional late-onset GCs (over 40 years old) [36,37] with more aggressive behavior, molecular heterogeneity and worse prognosis observed in EOGCs [38-41]. Thus, EOGCs may provide a unique clinical model to unravel the potential clinical impact of ARID1A mutations and deficiency on tumor onset in human cancers.
In this study, to address these unanswered key questions on the pathophysiology of ARID1A mutations and deficiency in human cancers, we undertook a comprehensive analysis of ARID1A genomic alterations, protein deficiency and associated alterations in tumor immune contexture in EOGC.
Materials and methods
Patient samples and tissue microarray
All patients included in this study had histologically confirmed GC who underwent surgical resection between January 2013 and February 2017 in Tongji Hospital. Informed consent was collected according to the Helsinki Declaration and ethical approval was granted by the ethical review committees at the Huazhong University of Science and Technology, Tongji Hospital. Overall survival (OS) was the interval from diagnosis to death, or to the date of the last contact. The Karnofsky score of all patients was evaluated to be at least 80. Of the 136 EOGC patients (≤40 years) eligible for this study, 20 were excluded because of insufficient tumor tissues, and 16 were excluded because of low DNA quality for sequencing, leaving 100 patients for the current analysis. Clinical information was obtained from the electronic medical records.
One tissue microarray (ST8018; Xi’an AiDi Biotechnology, Xi’an, China) of 40 young patients defined as EOGC (≤35 years) consisting of primary GC samples and matched peritumor tissues (PT) were purchased for immunohistochemistry (IHC) and multiplex immunohistochemistry (mIHC). The Characteristics of patients’ information is shown in Supplementary Table 7.
Targeted DNA sequencing
DNA was extracted from FFPE tissue blocks of 100 EOGC patients using QIAamp DNA FFPE Tissue Kit (QIAGEN, Catalog no. 56404) and the concentration of FFPE DNA samples was measured by Qubit dsDNA assay and FFPE DNA quality was then assessed to ensure A260/A280 is within the range of 1.8 to 2.0. All qualified DNA samples were sheared into 200 bp target size by sonication (Covaris, M220) for library construction by using NEBNext Direct Custom Panel v1.1 (#E7060B-X1AAJ, NEB, Ipswich, MA, USA), which was designed to enrich for DNA fragments of ARID1A gene for next-generation sequencing on the Illumina platform. Fragmentation of DNA was followed by denaturation, probe hybridization, adaptor ligation, adaptor cleaving and PCR amplification. Indexed samples were pooled and loaded onto flow cells for sequencing on a Hiseq Xten (Illumina, Inc., USA) according to the manufacturer’s protocol.
Sequencing data analysis
The mean sequencing depth of coverage for the ARID1A gene was more than 500× for all cases, with the exception of the non-targeted region in each case.
Sequence data was aligned to the reference human genome build GRCh37 (hg19) using Burrows-Wheeler aligner (BWA) 0.7.17-r1188. PCR duplication was marked by picard-2.20.3 before subsequent variant calling. GATK-4.1.4.0 Haplotype Caller was used to call variants. Variants with a coverage lower than 25 or a mutant allele frequency lower than 5% were filtered out. Variants were annotated by Ensembl-vep-release-97. Annotations were defined with ANNOVAR (http://annovar.openbioinformatics.org/en/latest). Population allele frequencies were extracted from the Exome Aggregation Consortium ExAC Browser (http://exac.broadinstitute.org/), 1000 Genomes (www.1000genomes.org), ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar), COSMIC database (https://cancer.sanger.ac.uk/cosmic), and the single-nucleotide polymorphism database of the National Center for Biotechnology Information (dbSNP), version 147 (www.ncbi.nlm.nih.gov/projects/SNP). We also compared our results with data from a large series of patients with ARID1A mutation in public TCGA database through cBioPortal (www.cbioportal.org).
IHC of ARID1A
IHC for detecting ARID1A was carried out on paraffin-embedded sections of 100 EOGC patients and another commercially purchased TMA. EOGC paraffin-embedded blocks were processed into 4 um thick sections and mounted on slides for staining. First, the sections and microarray slide were incubated at 60°C for 2.5 h, deparaffinized in xylene, hydrated in a gradient ethanol series (100%, 90%, 80%, 75%), incubated with 3% hydrogen peroxide to eliminate endogenous peroxidase activity, and subjected to antigen recovery by microwaving samples for 15 min in Tris-EDTA solution (pH=9.0). Sections were incubated with 5% BSA for 20 min at room temperature (27°C) to block nonspecific sites followed by primary antibody at 4°C overnight. Specific primary Antibody against ARID1A (1:1000, ab182560; Abcam, Cambridge, UK) was used for IHC. Next, sections were incubated with secondary Abs for 25 min at room temperature. After washing with TBST three times, slides were stained with 3,3-diaminobenzidine (DAB) and counterstained with hematoxylin and mounted with coverslips in DPX (Sigma, USA) for imaging. ARID1A immunoreactivity was detected in the nucleus, in both tumor and stromal cells, which were used as an internal positive control in all cases.
Evaluation of ARID1A expression
The ARID1A protein expression of 100 EOGC slides and commercially available TMA slide was evaluated for both intensity (0= negative, 1= weak staining; 2= moderate; 3= strong) and proportion of positively stained cells expressed as a percentage (0=0%; 1+ ≤10%; 2+ ≤11-50%; 3+ ≤51-80%; 4+ >80%. The intensity and proportion of stained cells were multiplied to produce the final score between 0 and 12 [42]. Patients were divided into low and high groups representing ARID1A protein expression below and above the median, respectively. Evaluation of ARID1A heterogeneous expression in EOGC patients was dependent on the proportion of absolutely negative cells in tumor cells. Low heterogeneous expression group was defined as having “<10% or >90% negative tumor cells”, while high heterogeneous group was characterized as containing 10%~90% negative tumor cells. Same evaluation was also conducted on gastric epithelial cells in peritumor tissues. Ten 200X magnification fields in each stained slice were randomly selected for observation and scoring. Sections were evaluated by two pathologists who were blinded to the clinical information, and disagreement was resolved by a third pathologist.
Seven-color immunohistochemical multiplex staining
Commercially available TMA slide (ST8018, Xi’an Alena Biotechnology Ltd., Co., Ltd., Xi’an, China) with 40 matched pairs of primary GC samples and peritumoral gastric mucosa tissues was stained by mIHC for PD-L1, CD8, CD3, CD4, Foxp3, and pan-Cytokeratin (Supplementary Figure 3).
TMA slide was deparaffinized and tissues were fixed with formaldehyde:methanol (1:10) prior to antigen retrieval in heated Citric Acid Buffer (pH 6.0) for 15 min microwave treatment. The slide was put through six sequential rounds of staining, each including a protein block with Antibody Diluent/Block buffer (ARD1001EA) followed by primary antibody and corresponding secondary horseradish peroxidase-conjugated polymer. Each horseradish peroxidase-conjugated polymer mediated the covalent binding of a different Opal fluorophore using tyramide signal amplification. This covalent reaction was followed by additional antigen retrieval in heated Citric Acid Buffer (pH 6.0) for 15 min to remove bound antibodies before the next step in the sequence. After all six sequential reactions, sections were counterstained with Spectra DAPI (FP1490A, PE) and mounted with Fluoromount-G fluorescence mounting medium (SouthernBiotech, UAB, USA).
Slide scanning and analysis for mIHC
Multiplex stained TMA slide was scanned using the Vectra 3.0 software (Perkin Elmer, Waltham, MA) and the resultant raw high-power fields (HPF) of each core were photographed for further analysis. Spectral unmixing, cell segmentation, and identification and quantification of cellular subpopulations of TMA cores were processed in InForm 2.1 Image Analysis software (Perkin Elmer) after spectral unmixing algorithm was determined on single-stained control of each marker in pre-experiment.
To determine the proximity between cells displaying distinct phenotypes within the same tissue section, we used the Spatial Analysis Module in HALO v2.0 digital image analysis software (Indica Labs, Corrales, NM), which is compatible with Vectra and InForm software [43-45]. The algorithm of this image analysis software was designed to calculate the number of cells within a given distance of another cell.
Statistical analysis
All statistical analyses were performed appropriately by using GraphPad Prism 7.0 (San Diego, California, USA) unless specified otherwise. Difference among groups with percentage data or quantitative data were compared using unpaired t-test and Mann-Whitney U test (two-tailed) respectively. Categorical data were tested using the chi-square test. For survival analyses, Kaplan-Meier plots were using Log-rank Mantel-Cox test. The Pearson’s correlation coefficient(r) was used for the correlation analyses between groups of T cell subpopulations. Univariate and multivariate analyses of the survival data were performed using the cox regression analysis in SPSS version 23 (Chicago, IL, USA). P values lower than 0.05 were considered as statistically significant.
Results
Analysis of ARID1A genomic alterations in early-onset gastric cancer (EOGC)
To catalog ARID1A genetic alterations in early-onset tumors, we performed deep targeted-sequencing with a high resolution coverage of the whole genomic region of ARID1A (including 3’- and 5’-untranslated regions, introns and exons) in EOGC patients (≤40 years, n=100). Among these samples, 33 wildtype tumors were identified without any ARID1A alteration and a total of 119 variations including 98 single-nucleotide variants (SNVs) and 21 insertion/deletion (INDELs) were found in 67 cases (Figure 1A and Supplementary Table 1). All variants detected in our study were heterozygous alterations. Thirty-three alterations in coding regions were observed in 27 samples. Intronic variants were found in 45 samples while alterations in 5’ and 3’UTR were found in 11 samples (Figure 1A). Interestingly, among 27 samples with ARID1A coding variants, 5 samples contained multi-coding variants and 14 samples exhibited one-coding variant (Figure 1B). Additionally, there were 11 patients, who had variants in both coding and noncoding regions (Figure 1A, 1B).
Figure 1.
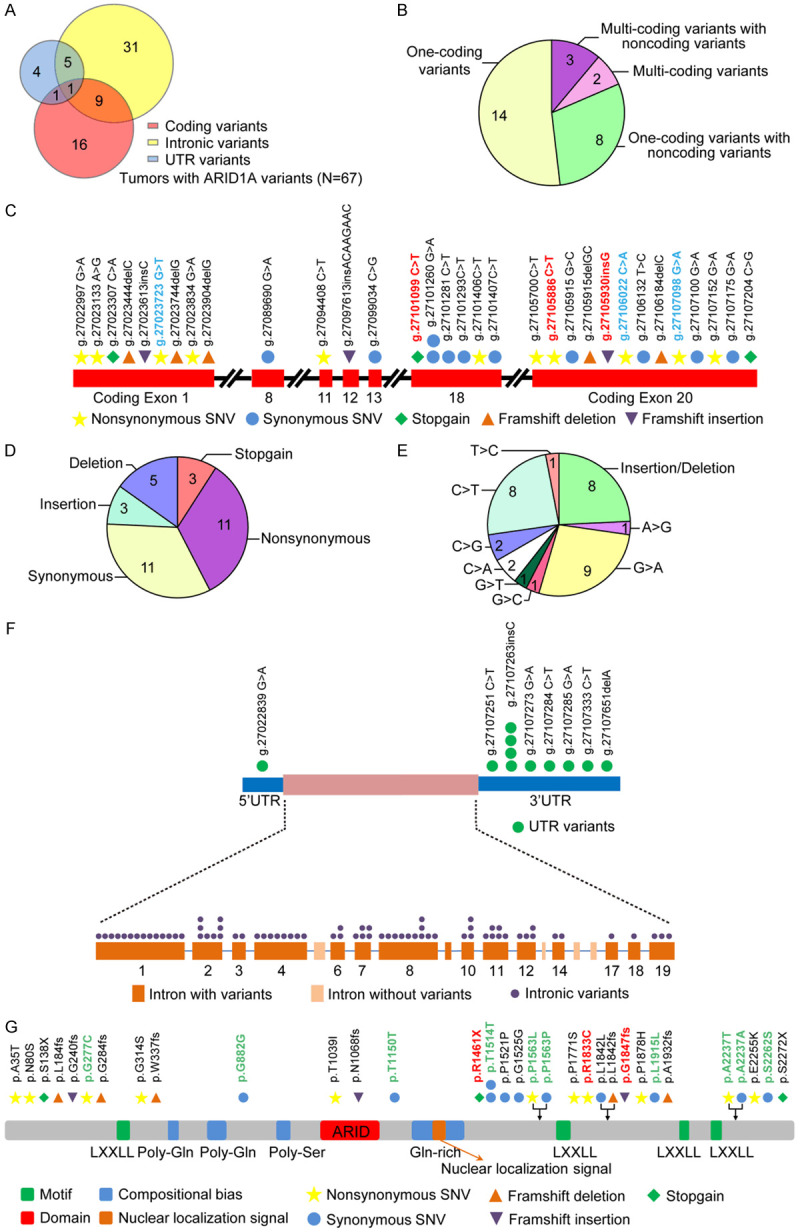
Genetic alterations dispersed in the coding and noncoding regions of ARID1A gene in EOGC tumors. A. Variants identified in different genomic regions of ARID1A (n=67). B. Multiple variants identified in the same tumor with ARID1A coding alterations (n=27). C. Schematic showing the feature and distribution of ARID1A variants in coding regions. (Red) Variants previously reported in TCGA; (Blue) Different variants at the same genomic sites previously identified in TCGA. Each symbol represents a variation event and variation types are depicted in different colors. D. The feature of 33 ARID1A coding variants identified in 27 tumors. E. The pattern of nucleotide-base change in 33 ARID1A coding variants identified in 27 tumors. F. Schematic showing the feature and distribution of ARID1A variants in noncoding regions. G. Schematic of ARID1A protein domains showing amino acid changes caused by ARID1A coding variants. (Red) Amino acid changes previously reported in TCGA; (Green) Different amino acid changes identified in the same sites previously reported in TCGA. UTR, un-translated region; SNV, single nucleotide variant.
We then plotted the disperse distribution of the 33 coding variants across the exon regions of ARID1A (Figure 1C). Surprisingly, the exonic variants were enriched in exons 1, 18 and 20. Among these 33 variants, there were 3 stop-gain variants, 8 insertion/deletion polymorphisms (INDELs), 11 synonymous SNVs and 11 nonsynonymous SNVs (Figure 1D). The nucleotide base change of coding variants showed a predominant pattern of C>T (8/33) and G>A (9/33) transition change (Figure 1E). The transition-transversion (ti/tv) ratio is 2.57 along with an insertion-deletion ratio of 0.6 (Figure 1D, 1E and Supplementary Table 1).
Next, we analyzed the distribution of variants in noncoding regions. In contrast to an enriched distribution pattern of coding variants, the noncoding variants showed a scattered distribution pattern (Figure 1F). Notably, among the noncoding regions with high frequency of variant allele occurrence, 14 variants (14/86), 11 variants (11/86) and 10 alterations (10/86) were located in intron 1, intron 8 and 3’UTR respectively (Figure 1F and Supplementary Table 1). Noncoding variants showed a dominant base change pattern of C>T (13/86) and G>A (16/86) similar to coding variants. However the base change of A>G was predominant found in noncoding variants (15/86 cases) compared to coding variants (1/27 cases) (Supplementary Figure 1). Furthermore, ti/tv ratio of 1.92 and the insertion-deletion ratio of 1.6 in noncoding regions were remarkably different from those of coding variants, suggesting that different mechanisms may underlie the molecular origins of variants located in different genomic regions (coding regions vs noncoding regions) (Supplementary Table 1).
To gain insight into potential pathogenic relevance of these variants, we then compared these coding variants with previously reported ARID1A SNVs in The Single Nucleotide Polymorphism Database (dbSNP) (Version 147) (Supplementary Table 1). We also annotated all 33 coding variants with the ExAC, 1000G (version 1000g2015aug) and ClinVar databases (Supplementary Table 1). Of 33 coding variants, 10 (30.3%) were archived in ExAC and 6 were archived in the 1000 Genome Project databases. Further, we found six out of 10 (60%) ExAC-archived variants showed a greater prevalence in East Asian populations than in the whole ExAC population. Interestingly, 4 out of the 6 variants archived in the 1000 Genome Project (66.7%) had a greater prevalence in East Asian populations than in the whole population. The other two variants were new variants identified from our study, which have not been reported in East Asian populations (Supplementary Table 1).
In the ClinVar database analysis, two out of the 33 coding variants (6%) were previously reported, whereas the pathogenic significance of the remaining 31 variants (94%) is unknown (Supplementary Table 1). We then analyzed the COSMIC database and found that 5 out of the 33 coding variants were archived. It was worthy of noting that 2 archived variants occurred concurrently in one patient case, that completely lost ARID1A protein expression (patient ID: G2-96) (Supplementary Table 1). Then we compared these 33 coding variants with ARID1A mutations identified in TCGA tumors (cBioPortal). We found 3 coding variants (1 nonsynonymous, 1 stop-gain and 1 frameshift insertion) were previously reported in TCGA databases (Figure 1C, 1G and Table 1). Interestingly, 3 variants (Figure 1C) were located at the same genomic site and 10 variants (Figure 1G) were located in same protein site but with different nucleotide/amino acid (AA) changes compared to TCGA mutations. Collectively these data revealed a spectrum of ARID1A variants with distinct features in both coding and noncoding regions in EOGC patients.
Table 1.
Variants reported in stomach adenocarcinoma (TCGA, Provisional)
| Sample ID | Sex | Age (Y) | Genome Location | Nucleotide Change | Amino Acid Change | Mutation type | Exon | avsnp147 | ExAC_ALL | ExAC_EAS | 1000g2015aug_all | 1000g2015aug_eas | Cosmic81_coding |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G2-6 | M | 24 | g.27105886 | c.5497C>T | p.R1833C | nonsynonymous SNV | 20 | rs372213935 | . | . | . | . | ID=COSM2235541; OCCURENCE=1 (stomach) |
| G2-47 | M | 40 | g.27101099 | c.4381C>T | p.R1461X | stopgain | 18 | NA | . | . | . | . | ID=COSM4031017; OCCURENCE=1 (oesophagus), 1 (stomach), 4 (endometrium), 1 (pancreas) |
| G2-96 | M | 37 | g.27105930 | c.5542dupG | p.G1847fs | frameshift insertion | 20 | NA | 2E-05 | 0 | . | . | ID=COSM1644335; OCCURENCE=3 (haematopoietic_and_lymphoid_tissue), 2 (large_intestine), 1 (lung), 1 (endometrium), 1 (salivary_gland), 1 (breast) |
NA, not available.
Correlation of ARID1A protein levels with ARID1A genomic alterations
Having identified a wide spectrum of genomic variants in both ARID1A coding and noncoding regions, next, we sought to determine whether these variants may have an impact on the levels of ARID1A protein. ARID1A protein expression was evaluated by immunohistochemistry staining (IHC) in this cohort of EOGC samples (n=100). Samples were divided into four groups based on the feature of ARID1A variants including coding variants, noncoding variants, 3’UTR variants and wildtype samples. We observed that the overall level of ARID1A protein was significantly reduced in all groups with ARID1A variants compared to the group with wildtype ARID1A (Figure 2A, 2B). Notably, tumors with 3’UTR variants showed a significant reduction of ARID1A protein to a similar degree as variants in coding regions (Figure 2B). Next, we analyzed different expression levels of ARID1A protein (high, low and negative expression) in tumors of each group (Figure 2C). As expected, the highest percentage of samples with negative ARID1A expression was found in the group with coding variants (22%, 6/27) and lowest in the group with noncoding variants (including UTR variants) (9.80%, 5/51). By contrast, 72.73% of cases (24/33) in the wildtype group and 41.18% of cases (21/51) in the noncoding variant group showed high-level ARID1A expression. Interestingly, we observed that 3’UTR variants had the highest percentage of cases containing low-expression ARID1A (80%, 8/10) with 1 case exhibiting high-level ARID1A expression and 1 case exhibiting negative ARID1A expression (Figure 2C). Surprisingly, although there was no negative ARID1A expression in the wildtype group, a significant portion of samples exhibited low ARID1A expression (27.23%, 9/33), while 14.81% of cases containing coding variants (4/27) exhibited high-level ARID1A expression. The specific variants with unexpected expression levels of ARID1A protein are summarized in Figure 2D. Among four patients with coding variants, who contained high-level ARID1A protein, one patient’s tumor surprisingly harbored three exon variants (Figure 2C, 2D). Together, these data indicated that ARID1A genomic variants in coding and 3’UTR regions had a significant impact on ARID1A protein levels, but were not the only molecular determinants of ARID1A protein levels in tumor cells. Next, we analyzed whether the presence of multiple ARID1A variants in a tumor may have an impact on the ARID1A protein level. We divided the samples into three groups based on the number of ARID1A variants found in the tumors including the wildtype group (no ARID1A variants), the one-site group (one variant) and the multi-site group (more than one variant) (Figure 2E). We found that tumors with one-site or multi-site variants exhibited reduced ARID1A protein levels compared to wildtype tumors (Figure 2F). Samples with multi-site variants had an increased percentage of negative ARID1A expression in tumors though, this was not statistically significant potentially due to the limited sample size in each group (Figure 2F). These results indicated that the genomic location and the number of ARID1A variants may alter protein expression in tumors.
Figure 2.
ARID1A variants affect its protein levels in EOGC tumors. A. Representative images of ARID1A protein analysis in tumors with indicated ARID1A variants (Coding, Noncoding, 3’UTR and Wildtype). ARID1A immunoreactivity was detected in the nucleus, in both malignant epithelial cells and stromal cells. ARID1A protein staining in stromal lymphocytes was served as an internal positive control. All scale bars equal 50 μm. B. IHC score of ARID1A expression in tumors with indicated ARID1A variants. Data were presented as mean ± s.d. p values were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. C. ARID1A protein levels in tumors with indicated ARID1A variants. D. ARID1A variants associated with unexpected protein expression. E. Samples stratified by the number of ARID1A variants identified in tumors. F. ARID1A protein levels in tumors with indicated number of ARID1A variants. p value was calculated by χ2 test.
Heterogeneous expression of ARID1A in EOGC
In addition to overall altered protein levels of ARID1A detected by IHC staining, we observed remarkable complexity of ARID1A protein expression patterns in both tumor and tumor-adjacent normal tissues. The presence of ARID1A heterogeneity in our study was determined by the obvious existence of negative staining in nuclei among neoplastic cells or normal epithelial cells in tumor cores and peritumor cores respectively. All sections were performed under strictly consistent condition optimized with verified antibody with very high specificity to avoid non-specific staining mentioned in Methods. Importantly, we carefully reviewed all stained sections and focused on the nucleus of tumor cells or normal epithelial cells to evaluate the heterogeneity.
As shown in Figure 3A, ARID1A expression levels could range from intense staining of all nuclei in tumor cells to the complete loss of staining with many intermediate staining phenotypes including the clonal type and the mixed type defined using the methods in previous publications [46,47]. This observation led us to further examine the association between ARID1A genomic variants and heterogenous expression of ARID1A protein. We divided samples into three groups based on ARID1A variants in coding, noncoding (including UTRs) and 3’UTR regions (Figure 3B). Nearly half of samples with ARID1A variants exhibited high heterogenous expression of ARID1A (Figure 3B). Surprisingly, samples containing 3’UTR variants showed the highest level of heterogeneous ARID1A expression among all variants (Figure 3B and 3C). There was no significant correlation between the extent of ARID1A heterogeneous expression and the number of concurrent variants in tumors (Figure 3D), while we observed samples with low ARID1A protein levels exhibited an increased heterogeneity in ARID1A expression compared to samples with high ARID1A expression (Figure 3E). In addition to heterogeneous ARID1A expression in tumor tissues, strikingly we observed localized or regional absence of ARID1A expression in the histologically normal gastric mucosa adjacent to tumor tissues in samples with ARID1A variants and also in samples with wildtype ARID1A (Figure 3F).
Figure 3.
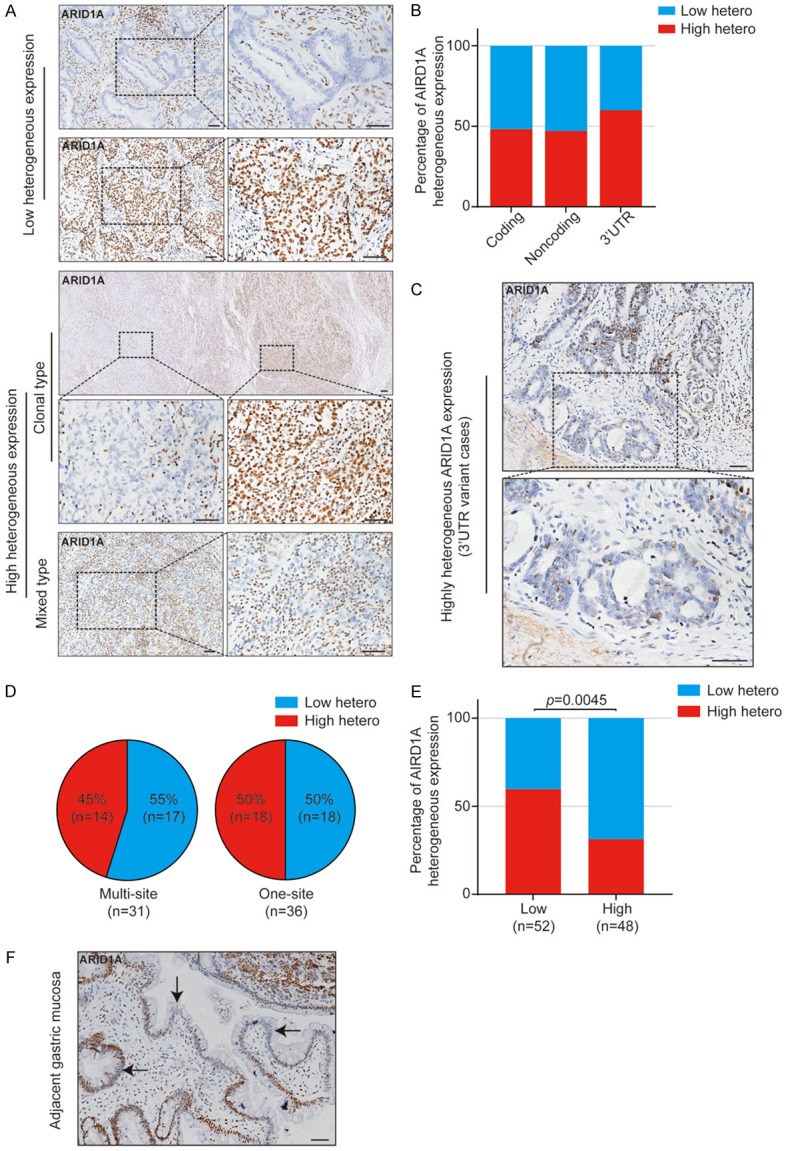
Heterogeneous expression of ARID1A protein in EOGC tumors. A. Representative images of different types of ARID1A heterogeneous expression. Low heterogeneous expression includes negative type and positive type. High heterogeneous expression includes clonal type and mixed type. B. ARID1A heterogeneous expression in tumors with indicated ARID1A variants. C. Representative images of ARID1A heterogeneous expression in tumors with 3’UTR variants. D. ARID1A heterogeneous expression in tumors with indicated number of ARID1A variants. E. The effect of ARID1A protein levels in tumors on its heterogenous expression. p value was calculated by χ2 test. F. Representative image of ARID1A heterogeneous expression in the histologically normal gastric mucosa tissue adjacent to the tumor tissue. Arrow heads indicate the ARID1A-deficient mucosa cells. All scale bars equal 50 μm.
Association of genomic alterations and heterogeneous expression of ARID1A with survival in EOGC patients
Next, we examined whether genetic alterations/heterogeneous expression of ARID1A associated with survival of EOGC patients. First, we grouped patients based on genomic regions of ARID1A variants or the number of variants identified in the tumors (Supplementary Figure 2A-G). Patients with ARID1A variants in tumors had a significantly longer survival than patients with wildtype ARID1A tumors (Supplementary Figure 2A). Patients with different types of ARID1A variants (coding, noncoding, intronic, 3’UTR, multi-site and one-site) had a longer survival trend compared to patients with wildtype ARID1A, although statistical significance was not achieved potentially due to the limited sample size in each sub-group (Supplementary Figure 2B-G).
We then grouped patients based on ARID1A protein levels. Patients with low expression (Supplementary Figure 2H) or with low heterogenous expression of ARID1A protein (Supplementary Figure 2I) had a longer survival trend compared to patients with high expression and with high heterogenous expression of ARID1A protein. Interestingly, in the group of patients with low expression of ARID1A in tumors, low ARID1A heterogenous expression was associated with a relatively improved survival compared to the high ARID1A heterogeneous expression (Supplementary Figure 2J). In contrast, in the group of patients with high expression of ARID1A in tumors, ARID1A expression heterogeneity did not show an association with survival (Supplementary Figure 2K). Consistent with this finding, low ARID1A heterogenous expression was associated with a trend of longer survival in patients with ARID1A variants (Supplementary Figure 2L) but not in patients with wildtype ARID1A (Supplementary Figure 2M). Among different types of variants, low ARID1A heterogenous expression showed a trend of improved survival in patients with ARID1A coding variants (Supplementary Figure 2N) and noncoding variants (Supplementary Figure 2O). Similar findings were also found in patients with multi-site variants (Supplementary Figure 2P) and patients with one-site ARID1A variants (Supplementary Figure 2Q). These data suggested that heterogeneity of ARID1A expression may have a more marked molecular impact on tumors with low ARID1A expression or with ARID1A genomic alterations.
Furthermore, we analyzed whether any clinical-pathological characteristics of EOGC patients were associated with ARID1A variants, ARID1A protein level and ARID1A heterogeneous expression, which might also contribute to patient survival (Supplementary Tables 2, 3, 4). We found that tumors with size bigger than 2 cm (P<0.0084), VEGF (P=0.0354) and thymidine phosphorylase (TP) positivity (P=0.0079) were significantly more frequent in patients with ARID1A coding variants than those in patients with wildtype ARID1A (Supplementary Table 2). In addition, VEGF negativity was significantly associated with patients with high ARID1A protein expression (P=0.0354) (Supplementary Table 3). No statistically significant results were found in patients stratified by ARID1A heterogenous expression (Supplementary Table 4). Additionally, multivariate Cox regression analysis with adjustments for characteristics including cigarette/alcohol consumption history, clinical stage, lymph node metastases, and Her-2/neu status revealed that ARID1A status remained as an independent association with survival in EOGC patients (HR 0.414, 95% CI 0.178-0.960, P<0.040) (Supplementary Tables 5, 6). Collectively, these results indicate that ARID1A variants, ARID1A protein levels or heterogeneous ARID1A expression in tumors may function as an independent molecular features prognosticating EOGC survival.
The protein level and heterogeneous expression of ARID1A in tumor tissues and peritumor normal tissues in EOGC
As shown in Figure 3F, we observed heterogeneous expression of ARID1A in tumor adjacent normal tissues. We thus systematically examined the level and heterogeneous expression of ARID1A protein in tumors (T) and in paired-adjacent histologically normal peritumor mucosa tissues (PT) using tissue microarrays (TMA) from an independent EOGC cohort (n=40; ≤35 years old). The clinical characteristics of the patients in the TMAs were described in Supplementary Table 7. Consistent with observations in the previous EOGC cohort (n=100), a striking heterogeneity of ARID1A expression in the T and PT tissues was found ranging from significant loss to high-level ARID1A expression in this cohort (Figure 4A-D). Of note, loss of ARID1A expression in PT tissues was not only found in samples with loss/low-expression of ARID1A in tumor cells, but also seen in samples with high-expression of ARID1A in tumor cells (Figure 4B). Based on the median expression level of ARID1A in T and PT tissues, we divided the samples into four groups including HH (high expression in both T and PT), HL (high expression in T/low expression in PT), LH (low expression in T/high expression in PT) and LL (low expression in T/low expression in PT) (Figure 4A-E). We also defined the samples by the level of heterogeneous ARID1A expression in T and PT tissues as shown in Figure 4F. Potentially due to limited sample size in each group, no statistical significance was detected when we examined the association among ARID1A protein levels or ARID1A heterogeneous expression in T and PT tissues (Figure 4E, 4F). However we indeed observed a potential molecular impact of ARID1A expression levels in T and PT tissues on the heterogeneous expression of ARID1A (Figure 4G-J). Samples with low ARID1A expression levels in tumor cells (T) showed an increased heterogeneity of ARID1A expression in T and also in PT tissues (Figure 4G, 4H). A similar pattern was also found in samples with low ARID1A expression detected in both T and PT tissues (Figure 4I, 4J). Strikingly, samples with high-ARID1A expression in T tissues but low-ARID1A expression in PT tissues (n=2) exhibited a dominant heterogeneity of ARID1A expression pattern in PT tissues, but not in T tissues, although the sample size was limited (Figure 4F and 4J). Nevertheless, these data suggested that ARID1A protein levels may change in both T and PT tissues, which potentially has a molecular impact on the heterogeneous expression of ARID1A not only in T tissues but also in PT tissues.
Figure 4.
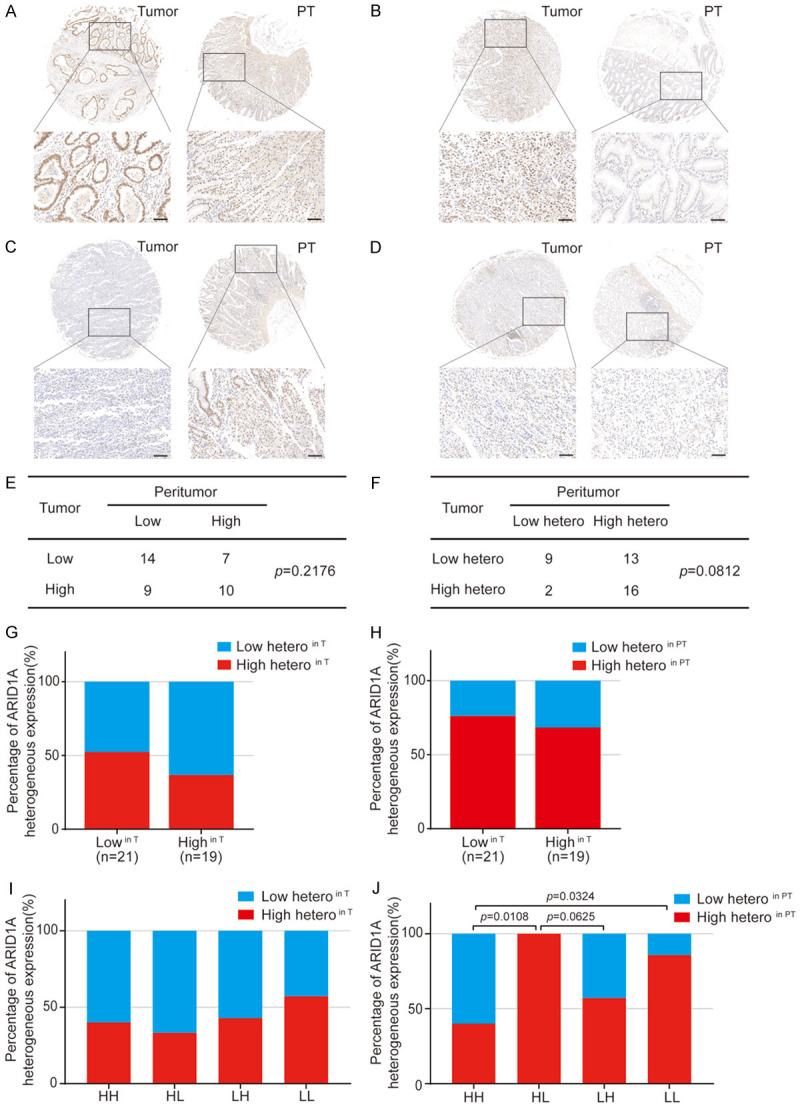
ARID1A protein analysis in tumor tissues and their paired normal peritumor tissues in EOGC. A-D. Representative images of ARID1A protein levels in tumor tissues (T) and peritumor tissues (PT). A. High in T and high in PT (HH); B. High in T and low in PT (HL); C. Low in T and high in PT (LH); D. Low in T and low in PT (LL). All scale bars equal 100 μm. E. The number of cases with indicated ARID1A protein levels in T and PT. p value was calculated by χ2 test. F. The number of cases with indicated ARID1A heterogeneous expression in T and PT. p value was calculated by χ2 test with Yates’ correction. G. ARID1A heterogenous expression in T in tumors stratified by high and low ARID1A protein levels in T. H. ARID1A heterogenous expression in PT in tumors stratified by high and low ARID1A protein levels in T. I. ARID1A heterogeneous expression in T in tumors stratified by high and low ARID1A protein levels in T and PT. J. ARID1A heterogenous expression in PT in tumors stratified by high and low ARID1A protein levels in T and PT. The p-value was calculated by Fisher’s exact test.
The effect of ARID1A-expression levels and heterogeneity on tumor-infiltrating T lymphocytes
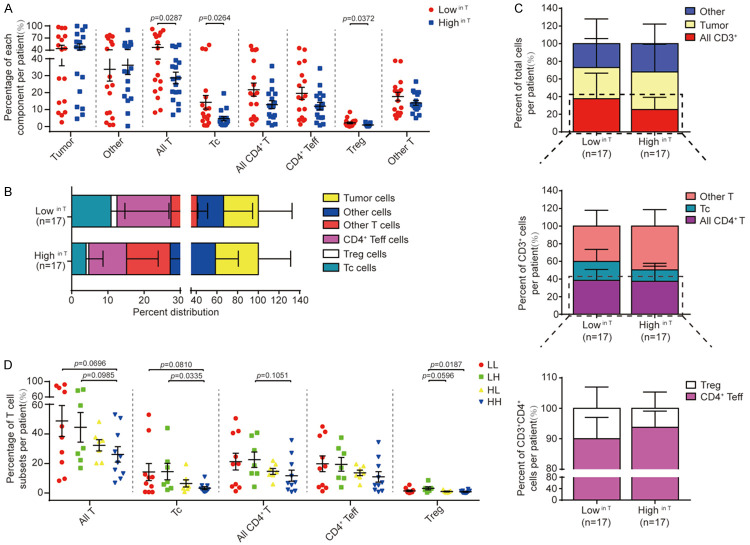
Studies from our group and others have reported that ARID1A deficiency in tumors is associated with an increased immune responsiveness, which is a significant contributor to patient survival. In the next step, we asked a previously unexplored question whether the level of ARID1A expression and/or the heterogeneous expression of ARID1A in tumor tissues and also peritumor normal tissues are associated with overall infiltration of T lymphocytes in tumors.
To answer this question, we examined the infiltration of various T lymphocyte subpopulations in tumors (T) of EOCG tissue microarrays (valid n=34, ≤35 years old) utilizing quantitative fluorescent multiplex immunohistochemistry staining (mIHC) (Supplementary Figure 3). As shown in Figure 5 and Supplementary Figure 4, in combination with histological properties such as cellular size and morphology, key markers CD3, CD8, CD4 and Foxp3 were examined to define T cell subpopulations including all T cells (CD3+), all CD4+ T cells (CD3+CD8-CD4+), cytotoxic T cells (Tc cells, CD3+CD8+CD4-), CD4+ effector T cells (CD4+ Teff, CD3+CD8-CD4+Foxp3-), regulatory T cells (Treg cells, CD3+CD8-CD4+Foxp3+) and ‘other T cells’ (any CD3+ cells negative for the three other markers). Pan-Cytokeratin and DAPI were used to identify epithelial cancer cells in tumor cores and nuclear stain respectively as previously described [48].
Figure 5.
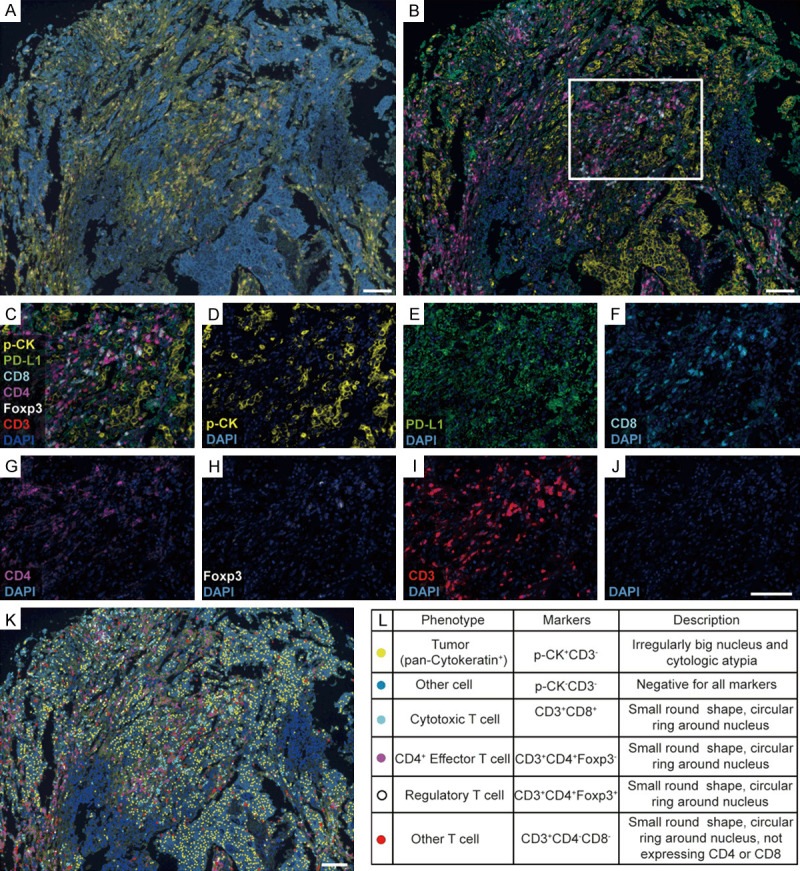
Multiplex immunohistochemistry (mIHC) analysis of T cell subpopulations in EOGC tumors. (A, B) Representative mIHC images of the tumor tissues: (A) raw image and (B) composite image after spectral unmixing. (C-J) Representative composite images showing staining markers after spectral unmixing: (C) all markers, (D) P-CK (cytoplasmic, visualized with Opal 520, pseudocolored yellow), (E) PD-L1 (membrane, Opal 540, pseudocolored green), (F) CD8 (membrane, Opal 570, pseudocolored cyan), (G) CD4 (membrane, Opal 620, pseudocolored megenta), (H) Foxp3 (nuclear, Opal 640, pseudocolored white), (I) CD3 (membrane, Opal 690, pseudocolored red) and (J) DAPI (nuclear, Spectral DAPI, pseudocolored blue). (K) Cell phenotype map identifying different cell subpopulations defined by the multiplex staining markers. (L) Summary of cell subpopulations, color codes and their associated markers. All scale bars equal 100 μm.
We first tested whether ARID1A expression in tumor cores (T) correlates with the alterations in TILs. We found that the percentage of overall T cells (namely CD3+ cells), Tc cells and Treg cells were significantly higher in patients with low ARID1A expression in tumor tissues (Lowin T) compared to those in patients with high ARID1A expression (Highin T) (Figure 6A-C), whereas the percentage of tumor cells (p-CK+CD3-), other cell components (p-CK-CD3-), overall CD4+ cells, CD4+ Teff cells and other T cells did not show a significant difference between these two groups (Figure 6A). Although ARID1A expression levels in PT tissues (Lowin PT/Highin PT) alone may not directly impact on T cell infiltration in tumors (Supplementary Figure 5A), low levels of ARID1A protein in both T and PT tissues (LL) were associated with a remarkable increase in the percentage of overall T cells (namely CD3+ cells), Tc cells and Treg cells among four groups of samples defined by ARID1A protein levels in T and PT (Figure 6D). As shown in Supplementary Figure 5B-D, the heterogeneity of ARID1A protein levels in T and PT did not exhibit a significant correlation with the overall number of TILs present in tumor tissues. Notably, Tc, Teff and Treg cells showed a positive correlation when they were compared in pairs (Supplementary Figure 5E-G) indicating a potential common mechanism in recruiting these T cell subpopulations. Collectively, these results indicated that ARID1A expression levels in both tumor and peritumor tissues inversely correlated with overall infiltration of a subpopulation of T cells including Tc cells and Treg cells, suggesting an active immune status in ARID1A-deficient EOGC.
Figure 6.
ARID1A protein levels correlate with the infiltration of T cell subpopulations in EOGC tumors. (A) The presence of indicated cell component in tumors stratified by the median ARID1A protein level in tumor tissues (lowin T vs highin T). (B) Relative distribution of cell components in tumors stratified by the median ARID1A protein level in tumor tissues (lowin T vs highin T). (C) Relative distribution of T cell subpopulations stratified by the median ARID1A protein level in tumor tissues (lowin T vs highin T). (D) The presence of indicated cell component in tumors stratified by the median ARID1A protein level in tumor and peritumor tissues (LL, LH, HL and HH) as defined in Figure 4. Statistical significance was computed by unpaired t-test. All data presented as mean ± s.d (B, C) or mean ± s.e.m (A, D).
The effect of ARID1A expression on the spatial organization and functioning density of tumor-infiltrating T cells
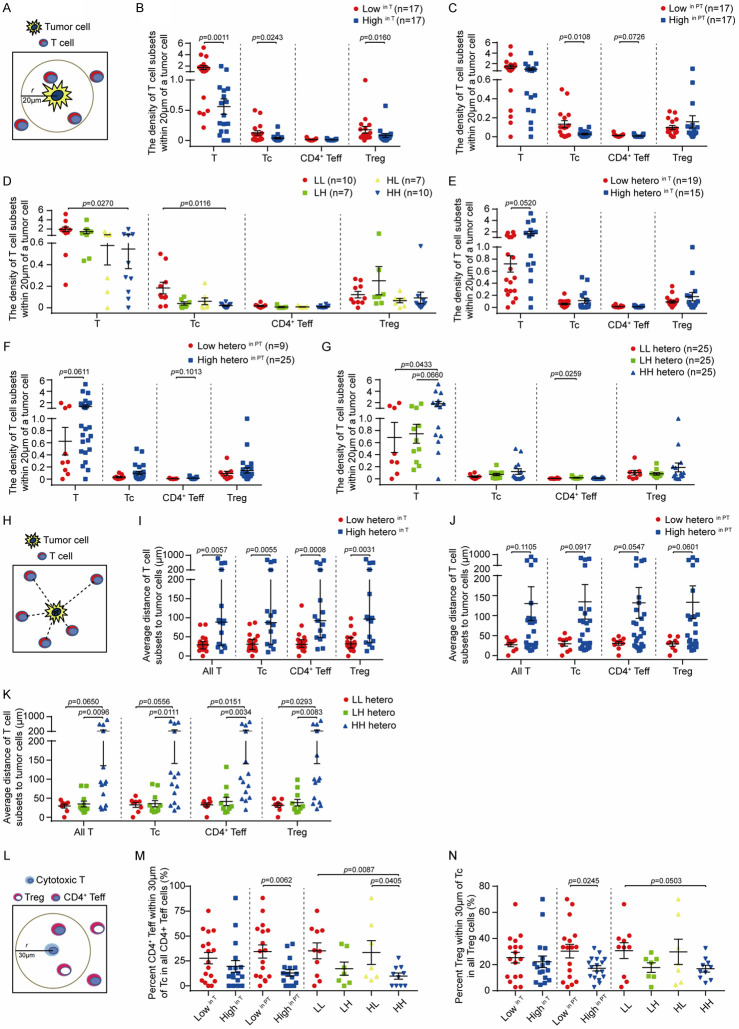
Recent studies have shown that not only the relative abundance, but also the spatial organization and functioning density of tumor-infiltrating T cells are important factors that determine anti-tumor immune responses [44]. Thus, we first tested whether the levels of ARID1A protein affects the spatial distribution of T cell subpopulations in the radius of 20 μm around tumor cells, which represents an enhanced probability for cell-cell contact (Figure 7A). We defined the functioning density of infiltrating T cells by calculating the number of T cells in each subpopulation relative to the number of tumor cells in the same region (Figure 7B-G). We found that the density of infiltrating total T cells, Tc cells and Treg cells was significantly higher in tumors with low ARID1A (lowin T) expression compared to tumors with high ARID1A expression (highin T) (Figure 7B). Interestingly, we also observed that tumors with low ARID1A expression in PT tissues (lowin PT) had a significant increase in the density of infiltrating Tc cells compared to tumors with high ARID1A expression in PT tissues (highin PT) (Figure 7C). Furthermore, samples with low ARID1A expression in both T and PT tissues (LL) showed a significant increase in the density of total T cells and Tc cells compared with other groups (Figure 7D). In contrast, a significant increase of Treg cell density was found in samples with ARID1A expression lowin T tissues and highin PT tissues (LH) (Figure 7D). Next, we tested whether the heterogenous expression of ARID1A in T and PT tissues may affect the density of T cell subpopulations in the radius of 20 μm around tumor cells. Due to the limited samples size (valid n=1) for HL hetero type (high heterogenous ARID1A expression in T and low heterogenous ARID1A expression in its paired PT), we only evaluated three subgroups of samples based on ARID1A heterogenous expression in T and PT tissues (LL hetero, LH hetero and HH hetero). As shown in Figure 7E-G, samples with high heterogenous ARID1A expression in T and PT (High heteroin T, High heteroin PT and HH hetero) showed an increased density of overall T cells. Furthermore, we analyzed the average distance of T cell subpopulations to tumor cells (Figure 7H). Strikingly, we observed high heterogenous ARID1A expression in T and PT tissues (High heteroin T or High heteroin PT and HH hetero) were associated with an increased average distance of T cell subpopulations to tumor cells, suggesting a reduced access of T cells to tumor cells (Figure 7I-K).
Figure 7.
ARID1A protein levels and heterogenous expression in tumor tissues and peritumor tissues impact on the density and spatial distribution of tumor-infiltrating T cell subpopulations. (A) Schematic of the density analysis of indicated T cell subpopulations within the area of 20 μm round a tumor cell. (B-G) The density of T cell subpopulations in two groups of tumors stratified by ARID1A protein characteristics in tumor tissues (T) and peritumor tissues (PT): (B) low and high ARID1A protein levels in T; (C) low and high ARID1A protein levels in PT; (D) low and high ARID1A protein levels in T and PT (HH, HL, LH and LL); (E) low and high ARID1A heterogenous expression in T; (F) low and high ARID1A heterogenous expression in PT; (G) low and high ARID1A heterogenous expression in T and PT. Statistical significance determined by Mann-Whitney U-test. (H) Schematic of the spatial distribution of T cell subpopulations as measured by their average shortest distance to tumor cells. (I-K) The spatial distribution of T cell subpopulations in groups of tumors stratified by ARID1A heterogeneous expression: (I) low and high heterogeneity in T; (J) low and high heterogeneity in PT; (K) low and high ARID1A heterogenous expression in T and PT. Statistical significance determined by Mann-Whitney U-test. (L) Schematic of the spatial distribution of CD4+ Teff and Treg cells within the area of 30 μm around cytotoxic cell (Tc) analyzed in m and n. (M) The distribution of CD4+ Teff cells in groups of tumors stratified by ARID1A protein levels in T, PT and both. (N) The distribution of Treg cells in groups of tumors stratified by ARID1A protein levels in T, PT and both. Statistical significance determined by unpaired t-test. All data presented as mean ± s.e.m.
Furthermore, we also analyzed the percentage of each T cell subpopulation (T cells, Tc cells, CD4+ Teff, Treg cells) recruited to the radius of 20 μm of tumor cells (Supplementary Figure 6A-G). Samples with low heterogeneous ARID1A expression in T or PT (Low heteroin T and Low heteroin PT) showed a higher percentage (Supplementary Figure 6B, 6C). Although the percentage of HH group was significantly lower than that of the other two groups, the percentage of LH hetero group was relatively higher than that of LL group (Supplementary Figure 6D). The similar results were also found in the comparison of proportion of T cell subsets (Tc cells, CD4+ Teff, Treg cells) adjacent to tumor cells in all Tc, CD4+ Teff and Treg cells respectively (Supplementary Figure 6E-G).
Among three T cell subpopulations including Tc, Teff and Treg cells, there was a strong positive correlation of presence of these cells in the proximal regions of tumor cells when two groups of them were compared (Supplementary Figure 6H-J). Given this concomitant recruitment of Teff and Treg cells with Tc cells to tumor microenvironment, we further tested whether ARID1A expression affects the spatial distribution of Teff and Treg cells in the proximal regions of Tc cells, which may regulate the function of Tc cells potentially through localized molecular interactions. A strong positive correlation of cell counts of Teff and Treg cells adjacent to Tc cells is shown in Supplementary Figure 6K. We further found that the percentage of Treg/Teff (CD4+) cells in a radius of 30 μm around Tc cells among all Treg/Teff (CD4+) cells that infiltrated in the tumor tissues was negatively associated with ARID1A expression in T tissue and PT tissues (Figure 7L-N). Taken together, these data indicated that the levels and heterogeneity of ARID1A expression in tumor tissues and also in peritumor tissues have a significant impact on the functioning density and spatial organization of T lymphocytes infiltrated into the tumors, as well as the spatial distribution of immunosuppressive Treg cells or supportive CD4+ Teff cells in proximity to cytotoxic T cells.
Discussion
In this study, we systematically characterized genetic and protein alterations of ARID1A in EOGC, a unique clinical model to unravel the genetic changes that are potentially associated with initiating stages of tumorigenesis and drive the onset of tumors. Results from our study discovered previously unreported molecular features of ARID1A alterations, which exhibit a significant impact on patient survival and tumor immune contexture.
ARID1A genetic variants and their associated-protein expression in EOGC tumors
ARID1A mutations were found in 30% of TCGA GCs, which were primarily identified by whole exome sequencing. Deep-targeted sequencing of ARID1A in our study enabled us to catalogue a full spectrum of ARID1A genomic alterations including the coding and noncoding regions. ARID1A variants were found in 67% EOGC samples (Figure 1). 27% EOGC cases contained variants in the coding regions, which is close to TCGA findings in late-onset GC. These data suggested a significant portion of tumors may contain ARID1A variants in noncoding regions. Among these ARID1A variants, 11 noncoding variants and 1 coding variant were identified as recurrent variants in more than 1 case, which have not been reported in TCGA datasets. Notably, the most frequently recurrent variant in 3’UTR (c.*16_*17insC) was found in 5 EOGC cases (5%), which was associated with reduced ARID1A protein expression. Compared to variants in other noncoding regions, variants in 3’UTR was remarkably associated with reduction of ARID1A protein expression levels to the same degree as variants in coding regions (Figure 2A, 2B). Several possible mechanisms may underlie the reduced protein expression levels resulting from noncoding genetic changes including impaired gene transcription, altered splicing process, or reduced mRNA stability [49,50]. It remains to be further determined how ARID1A 3’UTR variants may affect its protein expression. In addition, the EOGC cohorts used in this study were from a primarily Asian population, among whom the gastric cancer incidence and mortality rate are significantly higher than Western population [38-41]. It is of future research interests to compare the genetic alterations of ARID1A in EOGC patients with different ethnic origins. Additionally, one study using whole exome sequencing (WES) in EOGC and LOGC showed that ARID1A mutation rate was 14% in EOGC (median age 38 years) and 15% in late-onset GC (LOGC median age 67 years), suggesting there was no difference of ARID1A mutation rates between EOGC and LOGC [51]. In this study, we identified a variety of alterations in ARID1A noncoding regions, which were associated with reduced ARID1A protein expression. It remains to be examined whether similar genetic variations or a similar mutation rate might be observed in LOGC. It is worth mentioning that all ARID1A variants identified in our study were heterozygous variants. A similar phenomenon was found in one study of hepatocellular carcinomas, where nearly all of the ARID1A mutations were found to be heterozygous [13]. In addition, among tumors with identified ARID1A variants, 48% (31/67) of cases contained multiple ARID1A variants and exhibited a higher percentage of negative ARID1A protein expression. These data suggested that when a ARID1A variant occurs on one allele and ARID1A protein may express from the other allele, which likely leads to reduced levels of ARID1A expression and may mediate a functional haploinsufficiency in promoting tumorigenesis. When multiple variants occur in the same tumor, possibly on different alleles, it may dampen ARID1A expression from both alleles and thus lead to loss of ARID1A expression without homozygous genetic hits. Surprisingly, we observed that 27% (9/33) of tumors with wildtype ARID1A showed a reduction in ARID1A expression (Figure 2E, 2F). This result suggested that epigenetic changes such as promoter DNA methylation or protein stability control rather than genetic alterations may play an important role in regulating ARID1A protein levels in tumors.
The heterogeneity of ARID1A protein levels
While scoring ARID1A expression levels in the whole tissue sections of EOGC patients, we observed a variety of ARID1A protein levels in different regions of the same tumor samples (Figure 3A). The patterns of ARID1A expression detected by IHC staining ranged from lack of staining to strong staining of all nuclei in tumor cells with intermediate staining phenotypes. Our analysis further showed that compared to tumors with high ARID1A expression, tumors with low ARID1A expression exhibited a higher frequency of heterogeneous expression (Figures 3E and 4G). Interestingly, tumors with 3’UTR variants showed increased heterogeneity of ARID1A protein levels (Figure 3B, 3C). This result suggested that variants in noncoding regions may not completely abolish protein expression like nonsense mutations in the coding regions. However, variants in noncoding regions may modify protein levels by regulating mRNA stability, protein synthesis efficiency or protein stability, contributing to the variation in ARID1A expression. In addition to this possibility, the heterogeneity of ARID1A levels may also be caused by different molecular stages of clonal expansion during tumorigenesis, cell differentiation status or epigenetic modifications associated with subsets of tumor cells. Mechanisms underlying the heterogeneity of ARID1A levels remain to be further investigated.
The heterogeneity of ARID1A protein levels may lead to distinct molecular changes in tumor cells, which in turn may contribute to the biological phenotypes and clinical outcomes of tumors. Indeed, our analysis showed that the low heterogeneity of ARID1A protein levels was associated with an improved survival in EOGC patients (Supplementary Figure 2I-O). Furthermore, our study for the first time showed that heterogenous protein levels of ARID1A markedly affected the density and spatial distribution of TILs in tumors (Figure 7 and Supplementary Figures 5 and 6). High heterogeneous ARID1A levels were associated with an increased density of TILs in the proximal region of tumors cells (the radius of 20 μm around the tumor cells) (Figure 7E-G). Paradoxically, EOGC patients with high heterogeneous ARID1A levels exhibited a worse survival compared to patients with low heterogeneous ARID1A levels. To examine the potential mechanism underlying this unexpected observation, we further analyzed the spatial organization of TILs by measuring the average distance between TILs and tumor cells (Figure 7H-K). Interestingly, our data showed that tumors with high heterogenous ARID1A levels exhibited significantly increased distance between TILs and tumors cells. These data suggested that anti-tumor immune response resulting from increased density of TILs may be dampened by increased spatial distribution distance of TILs to tumor cells. Not only the density of TILs in tumors, but also the spatial organization of TILs around tumor cells needs to be examined in order to assess the impact of ARID1A protein levels on tumor immune responsiveness.
Heterogeneity and significance of ARID1A protein levels in peritumor tissues
Surprisingly, we unexpectedly observed that “partially loss of ARID1A” is a quite common phenomenon in adjacent gastric tissues among EOGC patients (Figure 3E). Previous studies already reported that the mutation status of ARID1A is a key event in the transformation from endometriosis to ovarian clear-cell carcinoma [9,52,53]. In addition, accumulation of somatic mutations have recently been found in normal colorectal epithelial cells, liver cells, and corresponding diseased cells, suggesting that mutations of “cancer-associated genes” could be involved in malignant transformation at the initiation of carcinogenesis [37,54-56].
Although the mechanisms underlying loss of ARID1A in peritumor normal tissues need to be further investigated, it is possible that a proportion of non-malignant gastric epithelial cells adjacent to tumor may already harbor ARID1A mutations or other regulatory factors affecting ARID1A expression.
Notably, our results showed that peritumor tissues more likely exhibited heterogeneous ARID1A protein levels in tumors with low ARID1A expression (Figure 4H), suggesting potential molecular interactions between loss of ARID1A in tumors and ARIDA protein levels in peritumor tissues. Indeed, we found the levels of ARID1A expression and heterogeneity in peritumor tissues were remarkably associated with the recruitment, the density and the spatial distribution of TILs in tumor tissues (Figure 7 and Supplementary Figure 6). These data indicated that the altered ARID1A expression in peritumor tissues may function as a major contributor to shape the immune microenvironment, which should be taken into consideration when ARID1A expression is used as a marker to analyze tumor immune responsiveness. In addition, these findings may provide a rationale to examine genetic alterations and protein deficiency of ARID1A in normal gastric mucosa adjacent to tumor tissue and in gastric premalignant lesions, which may provide unique insights into developing surveillance strategies to monitor tumor onset and progression.
ARID1A expression patterns as a predictive marker for immune responsiveness
For the first time, our study characterized the patterns of ARID1A protein levels in both tumor tissues and paired-adjacent histologically normal mucosa tissues (peritumor tissues) including the protein levels and the heterogeneity expression of ARID1A and their molecular impact on tumor immune contexture including the recruitment of TILs, the density of TILs around tumor cells and the spatial distribution of TILs (the distance) around tumor cells (Figures 6, 7 and Supplementary Figures 5 and 6). We found a significant inverse relationship between TILs (all T, Tc, Treg) levels and ARID1A protein levels. Interestingly, ARID1A protein levels in tumor-adjacent gastric mucosa had a remarkable influence on the density and the spatial distribution of TILs in tumor tissues. This observation indicated that ARID1A expression status in non-tumorous mucosa, as an integral component of the tumor microenvironment, exerts a significant molecular impact on shaping tumor immune contexture likely during the initiation and progression of normal mucosal cells to cancerous cells.
Previous studies have identified potential therapeutic strategies through targeting ARID1A deficiency in tumors. More recently, in a variety of cancer types, mutations and protein deficiency of ARID1A have been associated with an altered immune microenvironment in tumors and responses to immune checkpoint blockade [57-60]. Thus, it is a critical clinical need to develop optimal assessment of ARID1A expression status as a biomarker to stratify cancer patients for target therapy and immune therapy. Our data showed that a comprehensive evaluation of ARID1A expression patterns in tumor tissues and peritumor tissues provided novel insights into the impact of ARID1A alterations on the complexity of immune changes in tumors. Thus, in contrast to conventional analysis using the average strength of ARID1A immune staining in tumor cells, an evaluation of ARID1A expression patterns (levels and heterogeneity) in tumor tissues and peritumor tissues may represent a potential new strategy for better assessing the molecular impact of altered ARID1A protein levels and better stratifying patients before target and immune therapeutics.
Disclosure of conflict of interest
G.B.M. has received sponsored research support from Nanostring Center of Excellence, Ionis (Provision of tool compounds); has received clinical trials support from AstraZeneca, Genentech, GSK, Lilly; has ownership interest in Catena Pharmaceuticals, ImmunoMet, SignalChem, Tarveda; and is a consultant/advisory board member of AstraZeneca, Chrysallis Biotechnology, ImmunoMET, Ionis, Lilly, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Tarveda, Turbine, Zentalis Pharmaceuticals; has licensed technology for a homologous recombination deficiency assay to Myriad Genetics and digital spatial profiling to Nanostring. T.A.Y. has received sponsored research support (to Institution) from AstraZeneca, Bayer, Clovis, Constellation, Cyteir, Eli Lilly, EMD Serono, Forbius/Formation Biologics, F-Star, GlaxoSmithKline, Genentech, ImmuneSensor, Ipsen, Jounce, Karyopharm, Kyowa, Novartis, Pfizer, Ribon Therapeutics, Regeneron, Sanofi, Seattle Genetics, Tesaro, and Vertex Pharmaceuticals; And is a consultant/advisory board member of Almac, Aduro, AstraZeneca, Atrin, Axiom, Bayer, Calithera, Clovis, Cybrexa, EMD Serono, F-Star, Guidepoint, Ignyta, I-Mab, Jansen, Kyn Therapeutics, Merck, Pfizer, Roche, Seattle Genetics, and Zai Labs. G.P. has received sponsored research support from Pifzer. No potential conflicts of interest were disclosed by the other authors.
Supporting Information
References
- 1.Wu S, Fatkhutdinov N, Rosin L, Luppino JM, Iwasaki O, Tanizawa H, Tang HY, Kossenkov AV, Gardini A, Noma KI, Speicher DW, Joyce EF, Zhang R. ARID1A spatially partitions interphase chromosomes. Sci Adv. 2019;5:eaaw5294. doi: 10.1126/sciadv.aaw5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutta A, Sardiu M, Gogol M, Gilmore J, Zhang D, Florens L, Abmayr SM, Washburn MP, Workman JL. Composition and function of mutant Swi/Snf complexes. Cell Rep. 2017;18:2124–2134. doi: 10.1016/j.celrep.2017.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56–67. doi: 10.1016/j.gde.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong R, Liu L, Tian Y, Wang Y, Tian J, Zhu BB, Chen W, Qian JM, Zou L, Xiao M, Shen N, Yang H, Lou J, Qiu Q, Ke JT, Lu XH, Wang ZL, Song W, Zhang T, Li H, Wang L, Miao XP. Genetic variant in SWI/SNF complexes influences hepatocellular carcinoma risk: a new clue for the contribution of chromatin remodeling in carcinogenesis. Sci Rep. 2014;4:4147. doi: 10.1038/srep04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, Nagel ZD, Zou J, Wang C, Kapoor P, Ma X, Ma D, Liang J, Song S, Liu J, Samson LD, Ajani JA, Li GM, Liang H, Shen X, Mills GB, Peng G. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones S, Wang TL, Shih IeM, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih IeM, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, Lee SP, Ho SL, Chan AK, Cheng GH, Roberts PC, Rejto PA, Gibson NW, Pocalyko DJ, Mao M, Xu J, Leung SY. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 11.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, Zhu ZD, Zhou B, Liu XY, Liu RF, Fei QL, Chen H, Cai B, Zhou B, Xiao HS, Qin LX, Han ZG. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 13.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 15.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, Hruban RH, Eshleman JR, Iacobuzio-Donahue CA, Goggins M, Maitra A, Malek SN, Powell S, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, Badham N, Rafiq R, Brough R, Gulati A, Ryan CJ, Francis J, Vermulen PB, Reynolds AR, Reaper PM, Pollard JR, Ashworth A, Lord CJ. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun. 2016;7:13837. doi: 10.1038/ncomms13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukumoto T, Park PH, Wu S, Fatkhutdinov N, Karakashev S, Nacarelli T, Kossenkov AV, Speicher DW, Jean S, Zhang L, Wang TL, Shih IM, Conejo-Garcia JR, Bitler BG, Zhang R. Repurposing Pan-HDAC inhibitors for ARID1A-mutated ovarian cancer. Cell Rep. 2018;22:3393–3400. doi: 10.1016/j.celrep.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen J, Peng Y, Wei L, Zhang W, Yang L, Lan L, Kapoor P, Ju Z, Mo Q, Shih IeM, Uray IP, Wu X, Brown PH, Shen X, Mills GB, Peng G. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 2015;5:752–767. doi: 10.1158/2159-8290.CD-14-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Wang W, Zhang Y, Cieslik M, Guo J, Tan M, Green MD, Wang W, Lin H, Li W, Wei S, Zhou J, Li G, Jing X, Vatan L, Zhao L, Bitler B, Zhang R, Cho KR, Dou Y, Kryczek I, Chan TA, Huntsman D, Chinnaiyan AM, Zou W. Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J Clin Invest. 2020;130:2712–2726. doi: 10.1172/JCI134402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamura R, Kato S, Lee S, Jimenez RE, Sicklick JK, Kurzrock R. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J Immunother Cancer. 2020;8:e000438. doi: 10.1136/jitc-2019-000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y, Ichimura T, Ushiku T, Funahashi S, Tateishi K, Wada I, Shimizu N, Nomura S, Koike K, Seto Y, Fukayama M, Aburatani H, Ishikawa S. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 24.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, Chan KH, Chan AS, Tsui WY, Ho SL, Chan AK, Man JL, Foglizzo V, Ng MK, Chan AS, Ching YP, Cheng GH, Xie T, Fernandez J, Li VS, Clevers H, Rejto PA, Mao M, Leung SY. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 25.Wu RC, Wang TL, Shih IeM. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther. 2014;15:655–664. doi: 10.4161/cbt.28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werner HM, Berg A, Wik E, Birkeland E, Krakstad C, Kusonmano K, Petersen K, Kalland KH, Oyan AM, Akslen LA, Trovik J, Salvesen HB. ARID1A loss is prevalent in endometrial hyperplasia with atypia and low-grade endometrioid carcinomas. Mod Pathol. 2013;26:428–434. doi: 10.1038/modpathol.2012.174. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W, Awadallah A, Xin W. Loss of ARID1A/BAF250a expression in ovarian endometriosis and clear cell carcinoma. Int J Clin Exp Pathol. 2012;5:642–650. [PMC free article] [PubMed] [Google Scholar]
- 28.Anglesio MS, Papadopoulos N, Ayhan A, Nazeran TM, Noe M, Horlings HM, Lum A, Jones S, Senz J, Seckin T, Ho J, Wu RC, Lac V, Ogawa H, Tessier-Cloutier B, Alhassan R, Wang A, Wang Y, Cohen JD, Wong F, Hasanovic A, Orr N, Zhang M, Popoli M, McMahon W, Wood LD, Mattox A, Allaire C, Segars J, Williams C, Tomasetti C, Boyd N, Kinzler KW, Gilks CB, Diaz L, Wang TL, Vogelstein B, Yong PJ, Huntsman DG, Shih IM. Cancer-associated mutations in endometriosis without cancer. N Engl J Med. 2017;376:1835–1848. doi: 10.1056/NEJMoa1614814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayhan A, Mao TL, Seckin T, Wu CH, Guan B, Ogawa H, Futagami M, Mizukami H, Yokoyama Y, Kurman RJ, Shih IeM. Loss of ARID1A expression is an early molecular event in tumor progression from ovarian endometriotic cyst to clear cell and endometrioid carcinoma. Int J Gynecol Cancer. 2012;22:1310–1315. doi: 10.1097/IGC.0b013e31826b5dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang JZ, Li C, Liu XY, Hu TT, Fan ZS, Han ZG. Hepatocyte-specific arid1a deficiency initiates mouse steatohepatitis and hepatocellular carcinoma. PLoS One. 2015;10:e0143042. doi: 10.1371/journal.pone.0143042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, Roberts CW. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, Gopal P, Zhu M, Nassour I, Chuang JC, Maples T, Celen C, Nguyen LH, Wu L, Fu S, Li W, Hui L, Tian F, Ji Y, Zhang S, Sorouri M, Hwang TH, Letzig L, James L, Wang Z, Yopp AC, Singal AG, Zhu H. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell. 2017;32:574–589. e576. doi: 10.1016/j.ccell.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson MR, Reske JJ, Holladay J, Wilber GE, Rhodes M, Koeman J, Adams M, Johnson B, Su RW, Joshi NR, Patterson AL, Shen H, Leach RE, Teixeira JM, Fazleabas AT, Chandler RL. ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion. Nat Commun. 2019;10:3554. doi: 10.1038/s41467-019-11403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang SC, Nassour I, Xiao S, Zhang S, Luo X, Lee J, Li L, Sun X, Nguyen LH, Chuang JC, Peng L, Daigle S, Shen J, Zhu H. SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut. 2019;68:1259–1270. doi: 10.1136/gutjnl-2017-315490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kartha N, Shen L, Maskin C, Wallace M, Schimenti JC. The chromatin remodeling component Arid1a is a suppressor of spontaneous mammary tumors in mice. Genetics. 2016;203:1601–1611. doi: 10.1534/genetics.115.184879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giryes A, Oweira H, Mannhart M, Decker M, Abdel-Rahman O. Exploring the differences between early-onset gastric cancer and traditional-onset gastric cancer. J Gastrointest Oncol. 2018;9:1157–1163. doi: 10.21037/jgo.2018.06.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mun DG, Bhin J, Kim S, Kim H, Jung JH, Jung Y, Jang YE, Park JM, Kim H, Jung Y, Lee H, Bae J, Back S, Kim SJ, Kim J, Park H, Li H, Hwang KB, Park YS, Yook JH, Kim BS, Kwon SY, Ryu SW, Park DY, Jeon TY, Kim DH, Lee JH, Han SU, Song KS, Park D, Park JW, Rodriguez H, Kim J, Lee H, Kim KP, Yang EG, Kim HK, Paek E, Lee S, Lee SW, Hwang D. Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell. 2019;35:111–124. e110. doi: 10.1016/j.ccell.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Smith BR, Stabile BE. Extreme aggressiveness and lethality of gastric adenocarcinoma in the very young. Arch Surg. 2009;144:506–510. doi: 10.1001/archsurg.2009.77. [DOI] [PubMed] [Google Scholar]
- 39.Theuer CP, de Virgilio C, Keese G, French S, Arnell T, Tolmos J, Klein S, Powers W, Oh T, Stabile BE. Gastric adenocarcinoma in patients 40 years of age or younger. Am J Surg. 1996;172:473–476. doi: 10.1016/S0002-9610(96)00223-1. discussion 476-477. [DOI] [PubMed] [Google Scholar]
- 40.Dhobi MA, Wani KA, Parray FQ, Wani RA, Wani ML, Peer GQ, Abdullah S, Wani IA, Wani MA, Shah MA, Thakur N. Gastric cancer in young patients. Int J Surg Oncol. 2013;2013:981654. doi: 10.1155/2013/981654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strong VE, Russo A, Yoon SS, Brennan MF, Coit DG, Zheng CH, Li P, Huang CM. Comparison of young patients with gastric cancer in the United States and China. Ann Surg Oncol. 2017;24:3964–3971. doi: 10.1245/s10434-017-6073-2. [DOI] [PubMed] [Google Scholar]
- 42.Khalique S, Naidoo K, Attygalle AD, Kriplani D, Daley F, Lowe A, Campbell J, Jones T, Hubank M, Fenwick K, Matthews N, Rust AG, Lord CJ, Banerjee S, Natrajan R. Optimised ARID1A immunohistochemistry is an accurate predictor of ARID1A mutational status in gynaecological cancers. J Pathol Clin Res. 2018;4:154–166. doi: 10.1002/cjp2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, Green B, Soni A, Cuda JD, Stein JE, Sunshine JC, Succaria F, Xu H, Ogurtsova A, Danilova L, Church CD, Miller NJ, Fling S, Lundgren L, Ramchurren N, Yearley JH, Lipson EJ, Cheever M, Anders RA, Nghiem PT, Topalian SL, Taube JM. Multidimensional, quantitative assessment of PD-1/PD-L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab. J Immunother Cancer. 2018;6:99. doi: 10.1186/s40425-018-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Z, Bethmann D, Kappler M, Ballesteros-Merino C, Eckert A, Bell RB, Cheng A, Bui T, Leidner R, Urba WJ, Johnson K, Hoyt C, Bifulco CB, Bukur J, Wickenhauser C, Seliger B, Fox BA. Multiparametric immune profiling in HPV- oral squamous cell cancer. JCI Insight. 2017;2:e93652. doi: 10.1172/jci.insight.93652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gartrell RD, Marks DK, Hart TD, Li G, Davari DR, Wu A, Blake Z, Lu Y, Askin KN, Monod A, Esancy CL, Stack EC, Jia DT, Armenta PM, Fu Y, Izaki D, Taback B, Rabadan R, Kaufman HL, Drake CG, Horst BA, Saenger YM. Quantitative analysis of immune infiltrates in primary melanoma. Cancer Immunol Res. 2018;6:481–493. doi: 10.1158/2326-6066.CIR-17-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao TL, Ardighieri L, Ayhan A, Kuo KT, Wu CH, Wang TL, Shih IeM. Loss of ARID1A expression correlates with stages of tumor progression in uterine endometrioid carcinoma. Am J Surg Pathol. 2013;37:1342–1348. doi: 10.1097/PAS.0b013e3182889dc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samartzis EP, Samartzis N, Noske A, Fedier A, Caduff R, Dedes KJ, Fink D, Imesch P. Loss of ARID1A/BAF250a-expression in endometriosis: a biomarker for risk of carcinogenic transformation? Mod Pathol. 2012;25:885–892. doi: 10.1038/modpathol.2011.217. [DOI] [PubMed] [Google Scholar]
- 48.Jeuck TL, Wittekind C. Gastric carcinoma: stage migration by immunohistochemically detected lymph node micrometastases. Gastric Cancer. 2015;18:100–108. doi: 10.1007/s10120-014-0352-4. [DOI] [PubMed] [Google Scholar]
- 49.Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 50.van Ouwerkerk AF, Bosada FM, van Duijvenboden K, Hill MC, Montefiori LE, Scholman KT, Liu J, de Vries AAF, Boukens BJ, Ellinor PT, Goumans MJTH, Efimov IR, Nobrega MA, Barnett P, Martin JF, Christoffels VM. Identification of atrial fibrillation associated genes and functional non-coding variants. Nat Commun. 2019;10:4755. doi: 10.1038/s41467-019-12721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho SY, Park JW, Liu Y, Park YS, Kim JH, Yang H, Um H, Ko WR, Lee BI, Kwon SY, Ryu SW, Kwon CH, Park DY, Lee JH, Lee SI, Song KS, Hur H, Han SU, Chang H, Kim SJ, Kim BS, Yook JH, Yoo MW, Kim BS, Lee IS, Kook MC, Thiessen N, He A, Stewart C, Dunford A, Kim J, Shih J, Saksena G, Cherniack AD, Schumacher S, Weiner AT, Rosenberg M, Getz G, Yang EG, Ryu MH, Bass AJ, Kim HK. Sporadic early-onset diffuse gastric cancers have high frequency of somatic CDH1 alterations, but low frequency of somatic RHOA mutations compared with lateonset cancers. Gastroenterology. 2017;153:536–549. e526. doi: 10.1053/j.gastro.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samartzis EP, Noske A, Dedes KJ, Fink D, Imesch P. ARID1A mutations and PI3K/AKT pathway alterations in endometriosis and endometriosis-associated ovarian carcinomas. Int J Mol Sci. 2013;14:18824–18849. doi: 10.3390/ijms140918824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto S, Tsuda H, Takano M, Tamai S, Matsubara O. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod Pathol. 2012;25:615–624. doi: 10.1038/modpathol.2011.189. [DOI] [PubMed] [Google Scholar]
- 54.Brunner SF, Roberts ND, Wylie LA, Moore L, Aitken SJ, Davies SE, Sanders MA, Ellis P, Alder C, Hooks Y, Abascal F, Stratton MR, Martincorena I, Hoare M, Campbell PJ. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574:538–542. doi: 10.1038/s41586-019-1670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, Georgakopoulos N, Torrente F, Noorani A, Goddard M, Robinson P, Coorens THH, O’Neill L, Alder C, Wang J, Fitzgerald RC, Zilbauer M, Coleman N, Saeb-Parsy K, Martincorena I, Campbell PJ, Stratton MR. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574:532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 56.Shimizu T, Marusawa H, Matsumoto Y, Inuzuka T, Ikeda A, Fujii Y, Minamiguchi S, Miyamoto S, Kou T, Sakai Y, Crabtree JE, Chiba T. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology. 2014;147:407–417. e403. doi: 10.1053/j.gastro.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 57.Bitler BG, Fatkhutdinov N, Zhang R. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin Ther Targets. 2015;19:1419–1422. doi: 10.1517/14728222.2015.1062879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buglioni S, Melucci E, Sperati F, Pallocca M, Terrenato I, De Nicola F, Goeman F, Casini B, Amoreo CA, Gallo E, Diodoro MG, Pescarmona E, Vici P, Sergi D, Pizzuti L, Di Lauro L, Mazzotta M, Barba M, Fanciulli M, Vitale I, De Maria R, Ciliberto G, Maugeri-Sacca M. The clinical significance of PD-L1 in advanced gastric cancer is dependent on ARID1A mutations and ATM expression. Oncoimmunology. 2018;7:e1457602. doi: 10.1080/2162402X.2018.1457602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho HD, Lee JE, Jung HY, Oh MH, Lee JH, Jang SH, Kim KJ, Han SW, Kim SY, Kim HJ, Bae SB, Lee HJ. Loss of tumor suppressor ARID1A protein expression correlates with poor prognosis in patients with primary breast cancer. J Breast Cancer. 2015;18:339–346. doi: 10.4048/jbc.2015.18.4.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He F, Li J, Xu J, Zhang S, Xu Y, Zhao W, Yin Z, Wang X. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. J Exp Clin Cancer Res. 2015;34:47. doi: 10.1186/s13046-015-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.