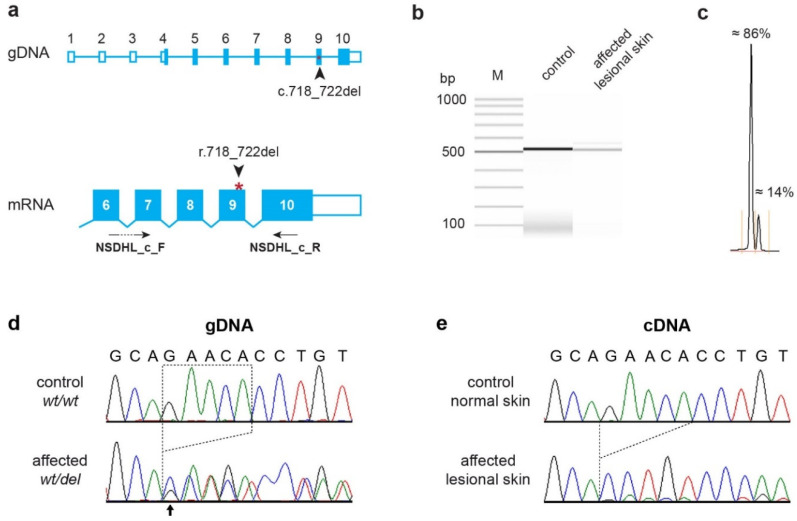
Figure 2.
Details of the NSDHL:c.718_722delGAACA variant. (a) Genomic organization of the canine NSDHL gene with 10 annotated exons. The position of the variant on the genomic DNA and the mRNA level are indicated. The position of the primers used for the RT-PCR experiment are also indicated. (b) RT-PCR amplification products obtained from cDNA from skin of a healthy control and lesional skin from the affected dog. The samples were analyzed on a FragmentAnalyzer capillary gel electrophoresis instrument. The predominant band in the affected dog corresponds to a correctly spliced transcript lacking 5 nucleotides of coding sequence (r.718_722del). The minor band that migrated slower than the main product most likely represents heteroduplex molecules consisting of one strand containing the 5-nucleotide deletion annealed to a wildtype strand. (c) Relative quantification of these two bands. Please note that the ratio of these end-point RT-PCR amplicons should only be seen as a semiquantitative proxy for the true ratio of transcripts. (d) Sanger electropherograms derived from genomic PCR products of a control dog and the affected dog. The five bases that are deleted in the affected animal are indicated with dotted lines. The arrow indicates the beginning of overlapping signals in the electropherogram of the affected dog. The intensities of the overlapping signals correspond well to the expected 50:50 allelic ratio for a heterozygous animal. (e) Analysis of mRNA. RT-PCR amplicons from the skin of a normal control dog and lesional skin of the affected dog were sequenced. The vast majority of NSDHL transcripts of the affected dog lacked 5 nucleotides corresponding to positions 718-722 in the coding sequence. Note the difference in relative signal intensity of the two alleles in the cDNA amplicon compared to the genomic sequence in the affected dog.