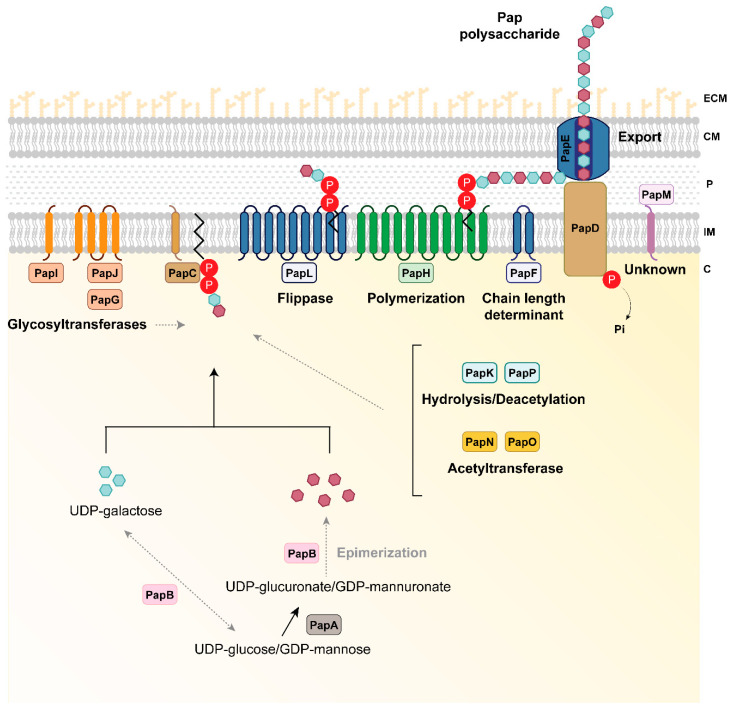
Figure 3.
Hypothetical model of P. fluorescens F113 Pap biosynthesis. PapA is a UDP-glucose/GDP-mannose dehydrogenase involved in the generation of the Pap polysaccharide residues (purple hexagons). PapB is predicted to be an epimerase that could be involved in the modification of the residues formed by PapA or providing the pool of sugar precursors for the synthesis of Pap. PapC is a phosphotransferase putatively involved in the phosphorylation of UDP-galactose in the synthesis of polysaccharide residues (blue hexagons) and the link of the sugar chain to a lipid carrier in the inner membrane. Once the constituting residues are synthesized, Pap is subjected to several modifications via glycosyltransferases (the cytoplasmic PapG and the inner membrane proteins PapI and PapJ), acetyltransferases (the cytoplasmic PapN and PapO), as well as deacetylation by the cytoplasmic PapK and PapP proteins to ultimately form the mature polysaccharide. The polymerization process and flippase machinery could be carried out by the inner membrane proteins PapH and PapL, respectively. The length of the synthesized polysaccharide could be controlled by the PapF inner membrane protein. The polysaccharide is exported to the milieu via the inner membrane tyrosine-kinase PapD and the outer membrane protein PapE. Whether the polysaccharide is secreted or remains attached to the cell-surface is unknown. There are additional proteins with unknown and no-predicted functions in the biosynthetic process such as PapM. Proteins are colored according to their predicted function as shown in Figure 1. Black solid lines indicate the steps catalyzed by each enzyme. Grey dashed lines indicate putative links or steps that need to be further investigated. C, cytoplasm; ECM, extracellular matrix; IM, inner membrane; P, periplasmic space; OM, outer membrane; P inside a red circle represents phosphate.