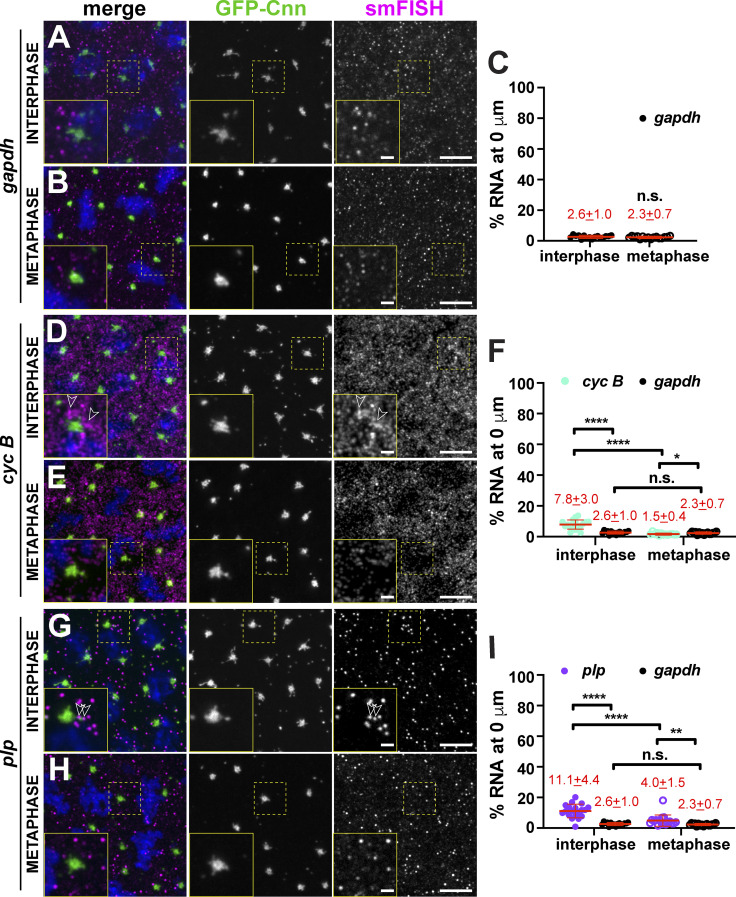
Figure 1.
Quantitative localization of mRNA to centrosomes. Maximum-intensity projections of smFISH (magenta) in NC 13 embryos expressing the centrosome marker GFP-Cnn (green). DAPI labels nuclei blue. Boxed regions enlarged in insets. Open arrowheads mark mRNA at the PCM. Quantification of the percentage of RNA overlapping with the centrosome surface (0 µm distance) is shown to the right, where each dot represents a single measurement from n = 16 interphase and metaphase (gapdh mRNA), 19 interphase and 13 metaphase (cyc B mRNA), and 19 interphase and 17 metaphase (plp mRNA) embryos, respectively. Mean ± SD displayed (red). (A–I) gapdh (A–C), cyc B (D–F), and plp mRNAs (G–I). Note that values for gapdh are reproduced from C for comparison. Table 1 lists the number of embryos, centrosomes, and RNA objects quantified per condition. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by ANOVA followed by Dunnett’s T3 multiple comparisons test. Scale bars: 5 µm; 1 µm (insets). n.s., not significant.