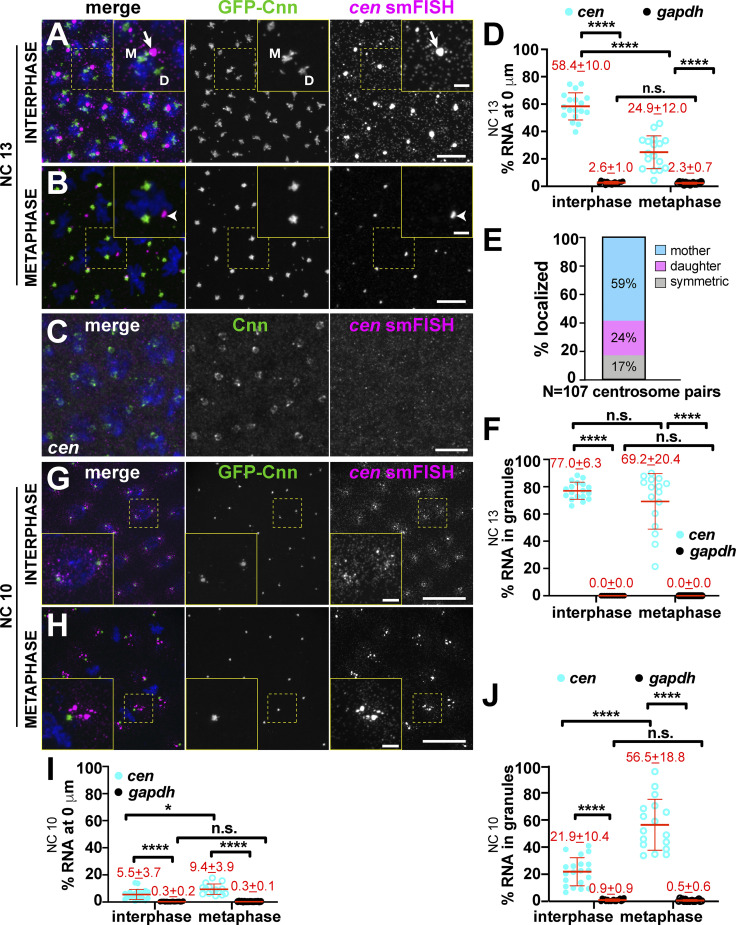
Figure 2.
cen mRNA localization to centrosomes is cell cycle regulated. Maximum-intensity projections of cen smFISH (magenta), where Cnn (green) labels centrosomes. Boxed regions enlarged in insets. (A) Interphase NC 13 embryo showing cen mRNA granules (arrow). Mother (M) and daughter (D) centrosomes noted. (B) Metaphase NC 13 embryo with cen mRNA (arrowhead) displaced from centrosomes. (C) cen mRNA is not detected in cen mutants. (D) Percentage of RNA overlapping with centrosomes in NC 13. Note that values for gapdh are reproduced from Fig. 1 C for comparison. (E) Frequency distribution of cen mRNA granule localization from n = 107 centrosome pairs and n = 5 embryos. (F) Percentage of cen mRNA residing within granules (≥4 overlapping RNA molecules) in NC 13. (G) Interphase NC 10 embryo with cen mRNA symmetrically distributed to centrosomes. (H) Metaphase NC 10 embryo with cen mRNA granules. (I) Percentage of RNA overlapping with centrosomes in NC 10. (J) Percentage of cen mRNA within granules in NC 10. Each dot represents a measurement from a single embryo. Mean ± SD displayed (red) from n = 16 interphase and metaphase (gapdh mRNA), 18 interphase and 17 metaphase (cen mRNA) NC 13; n = 19 interphase and 18 metaphase (gapdh mRNA); and 20 interphase and 17 metaphase (cen mRNA) NC 10 embryos, respectively. Table 1 lists number of objects quantified per condition. *, P < 0.05; ****, P < 0.0001 by ANOVA followed by Dunnett’s T3 multiple comparisons test (D, I, and J) and the Kruskal-Wallis test followed by Dunn’s multiple comparisons test (F). Scale bars: 10 µm; 2.5 µm (insets). n.s., not significant.