Abstract
MicroRNAs (miRs) carried in exosomes serve an important role in the pre-metastatic microenvironment and in intercellular interactions. However, the function of exosomal-miR-10a derived from primary colorectal cancer (CRC) cells on fibroblasts in the lung metastatic microenvironment of patients with CRC remains unclear. Reverse transcription-quantitative PCR was performed using samples from patients with CRC, and demonstrated that the expression levels of miR-10a were significantly lower in serum and cancer tissue samples from patients with CRC compared with in serum from healthy individuals and paired non-cancerous tissues, respectively. In addition, the expression levels of miR-10a were inversely associated with the invasion depth of CRC. Exosomal-miR-10a derived from CRC cells reduced the proliferative and migratory activities of primary normal human lung fibroblasts (NHLFs), and the expression levels of IL-6, IL-8 and IL-1β in NHLFs. The present study provided insight into the phenotypic alterations of NHLFs induced by exosomal-miR-10a derived from CRC cells, which may aid understanding of the mechanism underlying the process of CRC lung metastasis.
Keywords: colorectal cancer, exosome, microenvironment, lung fibroblasts, microRNA-10a
Introduction
The incidence of colorectal cancer (CRC) increased from 12.8 in 2003 to 16.8 per 100,000 in 2011 in China over the past few decades. Although the incidence of CRC is lower in China than that noted in other countries worldwide, the case-fatality and mortality/incidence ratios are higher (1). Overall, ~50% of patients with CRC may develop lymph node, liver, lung, peritoneum, brain and bone metastases; metastasis is associated with worse survival compared with that in patients without metastases (2). The liver is the most frequent target organ of CRC distant metastases, and ~1/3 of all patients with CRC present with or subsequently develop colorectal liver metastases (3). Only a limited number of patients with CRC present with lung metastasis. Although metastasectomy can uniformly improve cancer-specific survival in patients with liver metastases, it does not have the same effects for patients with lung metastasis and combined liver and lung metastases (4). To date, although several studies have focused on the liver metastasis of CRC, the mechanism underlying CRC lung metastasis is not fully understood.
The microenvironment has been recognized to serve an important role in tumor metastasis (5). Stromal cell reprogramming is a key factor in the remodeling of the internal microenvironment of tumors and in intercellular interactions (6). The stroma consists of fibroblasts, immune cells, blood lymphatic vessels and the extracellular matrix (ECM) (7). The complex composition of the microenvironment, notably its dynamic feature transformed by cancer cells, can enhance malignant progression. Mounting evidence has revealed that fibroblasts in normal tissues function as resting mesenchymal cells embedded within the interstitial fibrillar ECM, which can be activated to cancer-associated fibroblasts (CAFs) in a context-dependent manner during wound healing, tissue inflammation and organ fibrosis (8–10). Subsequently, CAFs can function synergistically with cancer cells to form an environment conducive to proliferation and metastasis (11).
It has been reported that tumor-derived exosomes are necessary for pre-metastatic microenvironment formation (12). Exosomes are a subset of extracellular vesicles (30–150 nm) that are released from cells upon fusion of an intermediate endocytic compartment, known as the multivesicular body, with the plasma membrane (13). They contain proteins, mRNA, microRNAs (miRNAs/miRs), small RNAs and/or DNA fragments that facilitate pre-metastatic niche formation by mediating communication between tumor cells and surrounding components, or by horizontally transferring their contents into recipient cells (14). miRNAs are small non-coding RNAs, which suppress gene expression at the post-transcriptional level via sequence-specific interactions with the 3′untranslated region of cognate mRNA targets (15). Previous studies have reported that exosomes contain a high level of miRNAs (16–18). These molecules have been shown to contribute to the development of metastasis in several types of cancer. For example, exosome-mediated transfer of miR-193a-3p, miR-210-3p and miR-5100 has been shown to promote metastasis of lung cancer cells by activating STAT3 signaling-induced epithelial-to-mesenchymal transition (EMT) (19). Tumor-derived exosomal-miR-1247-3p has also been shown to induce CAF activation and thus facilitate lung metastasis of liver cancer (20). However, the mechanisms underlying the activation of fibroblasts by CRC primary cells remain unclear, and are particularly obscure with regards to lung metastasis of CRC cancer.
Our previous study demonstrated that miR-10a expression was higher in primary CRC tissues compared with that noted in lymph node metastatic tissues. In addition, miR-10a was shown to suppress CRC metastasis to the liver by modulating EMT and the process of anoikis of CRC cells in vitro and in a nude mouse model (21). In the present study, the function of exosomal-miR-10a derived from CRC primary cells was investigated on a normal human lung fibroblast (NHLF) cell line established from the normal lung tissues. Initially, the expression levels of miR-10a were detected in serum and cancer tissues from patients with CRC, and the analysis aimed to assess whether miR-10a may be considered a blood-based biomarker for the diagnosis of patients with CRC. Subsequently, a NHLFs cell line was established from a patient with lung fibroma, and the ability of NHLFs to absorb exosomes from SW480 cells and the effects of exosomal-miR-10a on cell proliferation, migration and pro-inflammatory cytokine expression were assessed.
Materials and methods
Clinical specimens and cell lines
Human serum was collected from 20 healthy subjects and 40 patients with CRC who had not received chemotherapy or radiotherapy prior to radical resection at Tangshan People's Hospital (Tangshan, China). The patients with CRC were aged between 32 and 78 years old and included male and female patients, they were admitted to the Tangshan People's Hospital between April and October 2017. The CEA, CA-199 and CA-724 of the patients with CRC were detected by an electrochemical luminescence analyzer (E601; Roche Diagnostics) when they were admitted to the hospital. The TNM stage was determined according to The Eighth Edition AJCC Cancer Staging Manual (22). The healthy subjects were between 41 and 73 years old, which included male and female subjects, from the same hospital. Furthermore, 15 pairs of tissue samples from the 40 patients with CRC were collected (including adjacent normal tissues and cancer tissues). The use of clinical samples was approved by the Ethical Committee of Tangshan People's Hospital, and written informed consent was obtained from the individuals.
The NHLF cell line was established via the tissue block adherent method using tissues obtained from a 51-year-old male patient pathologically diagnosed with benign lung tumors in August 2017. The collection of a fresh clinical sample from this patient was approved by the Ethical Committee of Tangshan People's Hospital and written informed consent was obtained. Firstly, fresh normal lung tissue was collected >2.0-cm away from the lesion and rapidly added to Dulbecco's modified Eagle's F12 medium (DMEM/F12; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 0.1 mg/ml streptomycin. Within 20 min, the sample was washed twice with PBS, cut into 1.5–2.0-mm3 fragments and centrifuged at 200 × g for 5 min at 26°C. The small lung tissue fragments (1.5–2.0 mm3) were then collected into a cell culture flask filled with DMEM/F12 supplemented with 20% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin, and cultured at 37°C in a humidified incubator with 5% CO2 for 1 h. Subsequently, the culture flask was inverted for 24 h, after which the culture flask was placed in the humidified incubator and the culture medium was replaced with DMEM/F12 supplemented with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin. During the process of cell culture, DMEM/F12 was replaced every 3–4 days until cell confluence reached 80%. For the present study, passage 3–7 primary NHLFs were used.
The human colon cancer SW480 cell line was donated by Professor Hua Tang (Tianjin Medical University, Tianjin, China). The cells were conventionally cultured in minimum essential medium α (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin, and incubated at 37°C in a humidified incubator supplemented with 5% CO2.
Cell immunofluorescence
Third generation NHLFs were identified by cell immunofluorescence. Cells were seeded into 6-well plates at a density of 50% and incubated in a 37°C incubator containing 5% CO2 for 3–5 days until they had reached 80% confluence. Subsequently, cells were fixed with 2% paraformaldehyde in PBS for 10 min, permeabilized for 10 min in PBS-0.01 Triton X-100 (Sigma-Aldrich; Merck KGaA), and then blocked with 5% bovine serum albumin (cat. no. 0332-100G; Amresco LLC) to suppress non-specific reactions at room temperature for 60 min. Cells were then incubated with anti-cytokeratin-18 (CK-18) (1:500; cat. no. ab32118; Abcam) and anti-α-smooth muscle actin (α-SMA; 1:500; cat. no. SRP05217; Tianjiin Saierbio) overnight at 4°C, followed by incubation with Alexa Fluor® 555-conjugated secondary anti-rabbit antibody (1:500; cat. no. A-21-430; Invitrogen; Thermo Fisher Scientific, Inc.) for 60 min at room temperature in the dark. The negative isotype was incubated with PBS in the same conditions as the CK-18 and α-SMA antibodies. Finally, DAPI was used for nuclear staining at room temperature for 2 min. Cells were observed under an IX71 fluorescence microscope (Olympus Corporation).
Flow cytometry
For identifying primary cells, third generation NHLFs were cultured in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin in 6-well plates for 3–5 days, and 1×106 cells were harvested to detect α-SMA and CK-18 cell makers. Cells were blocked with blocking buffer (0.5% BSA in 1X PBS) after one wash, and were fixed with 2% paraformaldehyde at room temperature for 10 min. Subsequently, cells were washed once and permeabilized with 0.5 ml permeabilization solution (cat. no. 340973; BD Biosciences) at room temperature for 10 min. Cells were then washed once, incubated in blocking buffer for 30 min at room temperature, and incubated with α-SMA (1:20; cat. no. SRP05217; Tianjiin Saierbio) and CK-18 (1:20; cat. no. ab32118; Abcam) primary antibodies for 30 min at room temperature. Finally, cells were washed twice and incubated with Alexa Fluor® 555-conjugated secondary antibody (1:500) for 30 min at room temperature. Cells were resuspended in 1X PBS and analyzed using a BD FACSAria™ II (BD Biosciences) with BD FACSDiva™ software (BD Biosciences).
Extraction and identification of exosomes
SW480 cells at a density of 80% were cultured in cell culture flasks containing fresh DMEM/F-12 supplemented with exosome-depleted FBS. The cells were transfected with miR-10a mimics (5′-UACCCUGUAGAUCCGAAUUUGUGCAAAUUCGGAUCUACAGGGUAUU-3′) and negative control (NC) mimics (5′-UUCUCCGAACGUGUCACGUTT3ACGUGACACGUUCGGAGAATT-3′), purchased from Shanghai GenePharma Co., Ltd., at 37°C for 48 h, according to the manufacturer's protocol, using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, supernatants from the different groups were collected, centrifuged at 300 × g at 25°C for 10 min and filtered through 0.45-µM filters. Supernatants were again collected, and centrifuged at 2,000 × g for 10 min, 10,000 × g for 30 min and twice at 100,000 × g for 70 min using the Optima XPN-100 (Beckman Coulter, Inc.) all at room temperature. The exosome pellet was resuspended in PBS, ultra-centrifuged at 100,000 × g for 70 min, resuspended in 100 µl 1X PBS and stored at −80°C. Exosomes were observed under a JEM-1200EX transmission electron microscope (JEOL, Ltd.) and quantified using NanoSight LM10 (Malvern Instruments, Ltd.), which was conducted by Guangzhou EpiBiotek Co., Ltd..
RNA extraction and reverse transcription quantitative PCR (RT-qPCR)
The blood samples were collected from the patients with CRC, and then the serums were extracted from the blood at room temperature at 200 × g for 10 min within 30 min. Total RNA was extracted from tissues and serum using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA was tested for quality via 1% agarose gel electrophoresis. cDNA was synthesized using the PrimeScript™ 1st Strand cDNA Synthesis kit including DNase (Takara Bio., Inc.) from 3 µg total RNA with a thermocycler (Arktik™ 96; Thermo Fisher Scientific, Inc.) Stem-loop RT-qPCR (Suzhou GenePharma Co., Ltd.) analysis for the detection of miR-10a in tissues, cells and exosomes was conducted as previously described (21). The qPCR assay was performed as follows: 95°C for 3 min, and 40 cycles of 95°C for 12 sec and 62°C for 40 sec. The fold-change was analyzed using the 2−∆∆Cq method (23). Cel-miR-39 (Shanghai GenePharma Co., Ltd.) is a miRNA that is not expressed in human cells, so it was used as an external parameter for miR-10a in serum samples, and U6 was used as a housekeeping gene for miR-10a in tissues and cells.
TRIzol was used for RNA extraction from the NHLF cells. RT-qPCR analysis of IL-6, IL-8 and IL-1β was performed using PrimeScript™ 1st Strand cDNA Synthesis kit (Takara Bio, Inc.) and random primers for RT. The process of RT was performed at 65°C for 10 min, 25°C for 5 min, 0°C for 2 min, 42°C for 30 min, 70°C for 10 min. qPCR was conducted using SYBR Premix Ex Taq (Takara Bio, Inc.) and a PikoReal 96 RT-PCR system (Thermo Fisher Scientific, Inc.). qPCR was performed at 95°C for 4 min, then 33 cycles at 95°C for 1 min, 55°C for 1 min and 72°C for 1 min, and finally 72°C for 10 min. β-actin was used to normalize the expression levels of IL-6, IL-8 and IL-1β. The primer sequences are provided in Table I. All experiments were carried out at least in triplicate (24).
Table I.
Primer sequences used for reverse transcription-quantitative PCR.
| Gene | Primer sequence (5′-3′) |
|---|---|
| miR-10a | F: TGCGGTACCCTGTAGATCCG |
| R: CCAGTGCAGGGTCCGAGGT | |
| U6 | F: ATTGGAACGATACAGAGATT |
| R: GGAACGCTTCACGAATTTG | |
| Cel-miR-39 | UCACCGGGUGUAAAUCAGCUUG |
| IL-6 | F: ACTCACCTCTTCAGAACGAATTG |
| R: CCATCTTTGGAAGGTTCAGGTTG | |
| IL-8 | F: TTTTGCCAAGGAGTGCTAAAGA |
| R: AACCCTCTGCACCCAGTTTTC | |
| IL-1β | F: ATGATGGCTTATTACAGTGGCAA |
| R: GTCGGAGATTCGTAGCTGGA | |
| β-actin | F: ACTGTGCCCATCTACGAGG |
| R: GAAAGGGTGTAACGCAACTA |
miR-10a, microRNA-10a; NC, negative control; F, forward; R, reverse.
Trace experiment of exosomes
SW480 cells in the miR-10a mimics and NC groups were pretreated with 6.5 mmol/l DiO (Beyotime Institute of Biotechnology) at 37°C for 20 min, according to the manufacturer's instructions. Subsequently, the exosomes were collected from the SW480 supernatants as aforementioned. The obtained DiO-labeled exosomes were incubated with NHLFs pretreated with 6.5 mmol/l DiL (Beyotime Institute of Biotechnology) at 37°C for 10 min. The location of exosomes was observed using an IX71 fluorescence microscope (Olympus Corporation).
Proliferation assay
NHLFs (4×103/well) were seeded into 96-well plates, and equal quantities of exosomes from SW480 cells in the miR-10a mimics and NC groups were added into the wells with an exosome concentration of 10 µg/ml. To detect the proliferative ability of NHLFs, 10% CCK-8 (100 µl; Dojindo Molecular Technologies, Inc.) was added into the well and incubated with NHLFs at 37°C for 3 h. Absorbance was measured using a microplate reader at a wavelength of 450 nm (Multiskan FC; Thermo Fisher Scientific, Inc.). Each experiment was carried out in three replicate wells and was repeated three times.
Transwell migration assay
NHLFs were plated in 24-well Transwell plates at a density of 1×104/well in DMEM/F12 with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin. The medium in the upper chamber was free of FBS, whereas the medium in the lower chamber was supplemented with 10% FBS. For detecting exosome function, 6 µg SW480-derived exosomes were added to the inserts. After incubation at 37°C for 48 h, the cell inserts were fixed with 33% (v/v) acetic acid (glacial acetic acid: Methyl alcohol was 1:3) and stained with 0.1% crystal violet (Beijing Solarbio Science & Technology Co., Ltd.). Both the fixing and staining assays were conducted at room temperature for 10 min. Images of representative fields were captured with a light microscope, and the number of migrated cells per field was counted. The procedure was conducted as previously described (21), and the data were analyzed from three different wells per group.
Wound-healing assay
NHLFs at a density of 1×105 per well were plated into six-well plates in DMEM/F12 supplemented with 3% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin. The SW480-derived exosomes at a concentration of 10 µg/ml were added to each well at 37°C for 24 and 48 h. Subsequently, a wound was generated on the cell monolayers using a 200-µl pipette tip, and the NHLFs that migrated into the wounded area were observed under a light microscope at the specific time points. Wound closure was calculated with ImageJ software (version 1.46; National Institutes of Health).
Expression of inflammatory factors
To detect the effect of exosomes on the expression levels of cytokines, an equal number (1×106/flask) of NHLFs was plated in a 25 ml cell culture flask, and miR-10a exosomes and NC exosomes of the same quantities were added to the flasks. After incubation at 37°C for 48 h, the NHLFs in each group were collected and RT-qPCR was performed to detect the expression levels of the cytokines IL-6, IL-8 and IL-1β.
Statistical analysis
Data analysis was performed using SPSS software version 17.0 (SPSS, Inc.). Each experiment was carried out at least in triplicate and all results are presented as the mean ± SD. A paired Student's t-test was used to compare the paired tissue samples. Student's t-test was used to assess statistical significance. Significant association between miR-10a expression and clinicopathological parameters were assessed using the independent samples t-test. An ANOVA was performed to compare three groups followed by Scheffe post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-10a levels are expressed at lower levels in the serum and cancer tissues of patients with CRC
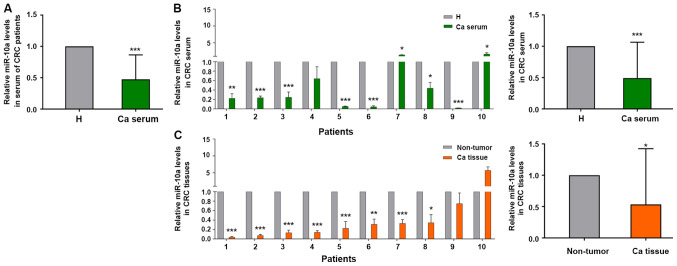
Our previous study discovered that miR-10a was highly expressed in primary CRC tissues compared with the corresponding expression in metastatic lymph node tissues (21). Therefore, the present study examined whether miR-10a was a biomarker for early diagnosis of patients with CRC. The expression levels of miR-10a were detected in mixed serum samples from 40 patients with CRC and 20 healthy subjects. The expression levels of miR-10a were lower in 32 out of the 40 serum samples from patients with CRC compared with those in the serum samples from the healthy subjects (not all 40 patient samples are shown; Fig. 1A). The expression levels of miR-10a in the serum samples from patients with CRC were negatively associated with the invasion depth of CRC; however, no significant association was noted between miR-10a expression and other clinicopathological indices (Table II).
Figure 1.
Relative expression levels of miR-10a in serum and cancer tissues from patients with CRC. (A) Reverse transcription-quantitative PCR analysis of miR-10a in serum samples from patients with CRC compared with serum from healthy subjects, nCRC=40, nH=20. (B) Expression levels of miR-10a in serum samples from randomly selected patients with CRC and healthy subjects, n=10. *P<0.05, **P<0.01, ***P<0.001 vs. H. (C) Expression levels of miR-10a in cancer tissues compared with adjacent non-tumor tissues from the same patients with CRC patient as (B), n=10. The expression of miR-10a in serum and tissues from patients with CRC was normalized to cel-miR-39 and U6 snRNA, respectively. The results are presented as the mean ± SD. All experiments were performed at least in triplicate, and Student's t-test was used to analyze the data. *P<0.05, **P<0.01, ***P<0.001 vs. non-tumor tissues. Ca, patients with CRC; H, healthy subjects; CRC, colorectal cancer; miR-10a, microRNA-10a.
Table II.
Association between miR-10a serum expression and clinicopathological features of patients with colorectal cancer.
| Variables | N (40) | miR-10a expression | P-valuesa |
|---|---|---|---|
| Age, years | 0.861 | ||
| ≤65b | 20 | 0.49±0.44 | |
| >65 | 20 | 0.46±0.35 | |
| Sex | 0.172 | ||
| Male | 23 | 0.42±0.40 | |
| Female | 17 | 0.57±0.34 | |
| Tumor sitec | 0.978 | ||
| Colon | 20 | 0.48±0.43 | |
| Rectum | 20 | 0.47±0.36 | |
| Tumor type | 0.372 | ||
| Adenocarcinoma | 37 | 0.49±0.39 | |
| Mucinouse | 3 | 0.37±0.28 | |
| Invasion depth | 0.039d | ||
| T1+T2 | 11 | 0.30±0.26 | |
| T3+T4 | 29 | 0.58±0.37 | |
| No. of positive nodes | 0.558 | ||
| 0 | 29 | 0.41±0.33 | |
| 1-3 | 6 | 0.53±0.45 | |
| 3+ | 5 | 0.65±0.55 | |
| Distant metastasis | 0.095 | ||
| M0 | 38 | 0.45±0.39 | |
| M1 | 2 | 0.92±0.16 | |
| TNM stage | 0.301 | ||
| I+II | 27 | 0.43±0.38 | |
| III+IV | 13 | 0.57±0.42 | |
| CEA, ng/ml | 0.593 | ||
| 0-5 | 22 | 0.44±0.33 | |
| >5 | 18 | 0.51±0.46 | |
| CA-199, U/ml | 0.611 | ||
| 0-39 | 36 | 0.46±0.39 | |
| >39 | 4 | 0.57±0.45 | |
| CA-724, U/ml | 0.260 | ||
| 0-6.9 | 37 | 0.49±0.39 | |
| >6.9 | 3 | 0.27±0.23 |
Student's t-test was used to compare two groups and ANOVA was used to compare three groups
Median age at operation
Proximal colon tumors are those arising in the cecum, ascending colon, hepatic flexure, or transverse colon; distal colon tumors are those arising in the splenic flexure, descending colon, or sigmoid colon; and rectal tumors are those arising in the rectosigmoid or rectum.
Statistically significant (P<0.05).
Mucinous type includes mucinous adenocarcinoma and signet ring cell carcinoma. CEA, carcinoembryonic antigen; CA-199, carbohydrate antigen 19-9; CA-724, carbohydrate antigen 72-4; miR-10a, microRNA-10a.
A total of 15 pairs of tissue samples were selected from the 40 patient samples, in order to detect the expression levels of miR-10a. After carefully checking the integrity of the tissue RNA, only 10 cases from these samples could be used for further study (Fig. 1B). The expression levels of miR-10a were detected in serum and tissue samples. The expression levels of miR-10a were lower in the serum samples from patients with CRC compared with those detected in the healthy subjects (Fig. 1B). A total of eight patients out of 10 exhibited lower miR-10a expression in cancer tissues compared with those noted in the adjacent normal tissues, which was consistent with the results obtained from serum samples (Fig. 1C). These results indicated that miR-10a expression was lower in serum and cancer tissue samples from patients with CRC, thus suggesting that this miRNA may be a potential biomarker for the diagnosis of CRC.
Establishment and identification of NHLFs
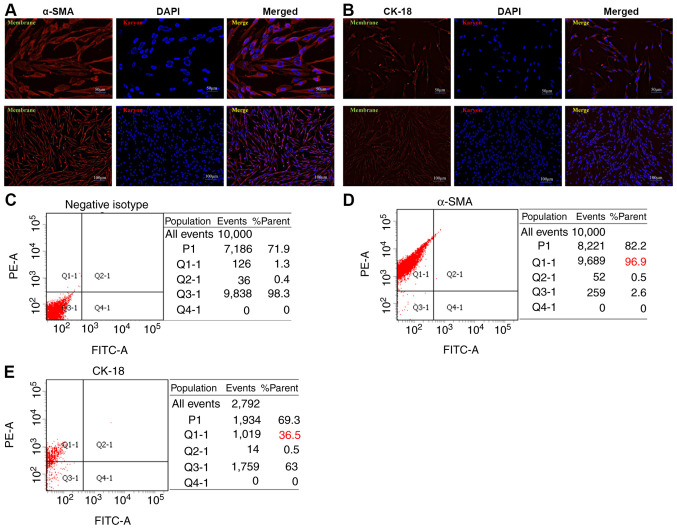
Fibroblasts have been recognized to be the dominant component of the tumor stroma. Previous studies have suggested a prominent functional role for these cells in cancer progression and metastasis (25–27). In the present study, NHLFs were established using the tissue block adherent method and the third generation of NHLFs was identified. The NHLFs exhibited classic spindle-shape morphology with a potential for planar polarity (Fig. 2A and B). α-SMA and CK-18 are the classic makers used for human fibroblast identification. The expression levels of α-SMA and CK-18 were examined in NHLFs by cellular immunofluorescence assays. α-SMA was expressed mainly in the cytoplasm and cellular membrane, whereas it was weakly expressed in the cell nucleus of NHLFs (Fig. 2A). CK-18 was weakly expressed in the cytoplasm, cellular membrane and cell nucleus of NHLFs (Fig. 2B). Consistent with the aforementioned results, flow cytometry indicated that the ratios of negative isotype (Fig. 2C), α-SMA (Fig. 2D) and CK-18 (Fig. 2E) expressed in NHLFs were 96.9 and 36.5%, respectively. Since low expression of CK-18 in NHLFs is a known marker for human epithelial cells (28,29), the expression analysis of these markers indicated that primary NHLFs were successfully established.
Figure 2.
Establishment and identification of NHLFs. Immunofluorescence analysis of (A) α-SMA and (B) CK-18 expressed in NHLFs. Sections of NHLFs were stained with α-SMA and CK-18 antibodies. Antigen-antibody complexes were visualized using DAPI and observed under a fluorescence microscope (top panel: magnification, ×200; bottom panel: magnification, ×400). Flow cytometric analysis results of (C) negative isotype, (D) α-SMA and (E) CK-18 in NHLFs. The experiments were performed at least in triplicate. NHLFs, normal human lung fibroblasts; α-SMA, α-smooth muscle actin; CK-18, cytokeratin-18.
Identification and tracer technique of exosomes from SW480 cells
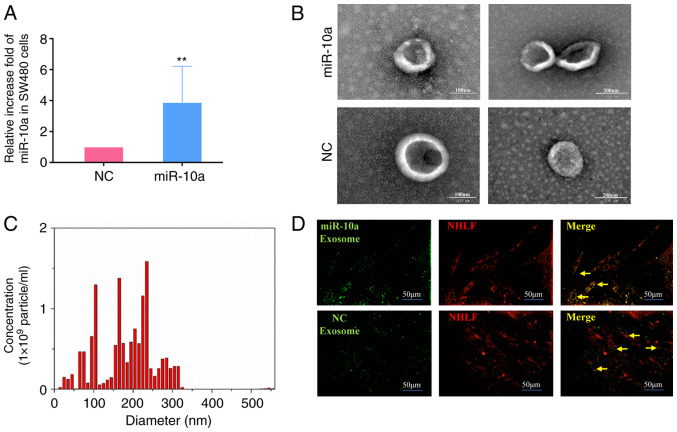
To further investigate the effects of miR-10a derived from SW480 cell exosomes, miR-10a was overexpressed in these cells by transfection with miR-10a mimics. Subsequently, exosomes were isolated. RT-qPCR indicated that the expression levels of miR-10a were significantly increased in exosomes from transfected cells compared with those noted in control subjects (Fig. 3A). Subsequently, electron microscopy and NanoSight equipment were employed to identify the shape and size distribution of the exosomes secreted from SW480 cells. The exosomes exhibited a hemispherical shape (Fig. 3B and C). The diameters of all exosomes were mainly concentrated at 0–300 nm and the majority of them were centered at 100–200 nm. Furthermore, it was shown that exosomes released by SW480 cells could be absorbed by NHLFs (Fig. 3D). Taken together, these data suggested that exosomes carrying miR-10a could form an important tool for communication between primary CRC cells and NHLFs.
Figure 3.
Identification and tracer analysis of exosomes from SW480 cells. (A) Expression of miR-10a in SW480 cells transfected with mimics-miR-10a (miR-10a) and NC; the isolated exosomes were detected by reverse transcription-quantitative PCR. U6 snRNA was used as an endogenous control. (B) Exosomes released by different groups of SW480 cells, as indicated, were detected by electron microscopy (left panel: magnification, ×100; right panel: magnification, ×200). (C) NanoSight particle diameter analysis of exosomes released by SW480 cells. (D) Fluorescence imaging of the delivery of DiO-labeled exosomes (green) to DiL-labeled NHLFs (red). Yellow arrows represent delivered exosomes. Representative images are presented (magnification, ×200). The experiments were performed at least in triplicate. **P<0.01 vs. NC. NHLFs, normal human lung fibroblasts; miR-10a, microRNA-10a; NC, negative control.
Exosomal-miR-10a from SW480 cells reduces NHLF proliferation, migration, and IL-6, IL-8 and IL-1β expression
In order to investigate the effect of exosomal-miR-10a from the primary CRC cells on fibroblasts in lung metastatic loci, the effects of exosomes derived from miR-10a-overexpressing SW480 cells were examined on NHLF features, including cell proliferation, migration and the expression of pro-inflammatory cytokines. NHLFs were respectively treated with the same quantity of exosomes from the miR-10a mimics and NC groups. The proliferative activity of the miR-10a group NHLFs was significantly decreased compared with that of the NC group within the first 2 days (Fig. 4A and D). In addition, wound-healing assays indicated that NHLFs exhibited reduced migratory activity when treated with exosomes overexpressing miR-10a at 24 and 48 h (Fig. 4B and E). The Transwell migration assay indicated similar results as those noted from the wound-healing assay at 48 h (Fig. 4C and F). In order to determine the effects of exosomes derived from miR-10a-overexpressing SW480 cells on the expression of inflammatory cytokines in NHLFs, the mRNA expression levels of IL-6, IL-8 and IL-1β were detected. The results demonstrated that all three inflammatory cytokines exhibited decreased expression levels in the miR-10a-overexpressing group (Fig. 4G). These data suggested that exosomal-miR-10a from SW480 cells may not only reduce NHLF proliferation and migration, but could also inhibit the mRNA expression levels of IL-6, IL-8 and IL-1β.
Figure 4.
Effect of exosomes derived from SW480 cells with miR-10a overexpression on NHLFs. (A and D) CCK-8 assay of NHLFs treated with the same quantities of exosomes derived from SW480 cells transfected with miR-10a mimics or NC. (B and E) Wound-healing assay of NHLFs treated with equal quantities of exosomes derived from SW480 cells transfected with miR-10a mimics or NC (magnification, ×40). (C and F) Migration assay of NHLFs treated with exosomes derived from SW480 cells in the indicated groups at 48 h. Migrated cells were counted and representative images are shown (magnification, ×40). The data from at least three wells per group were analyzed. (G) Reverse transcription-quantitative PCR analysis of IL-6, IL-8 and IL-1β expression in NHLFs treated with exosomes derived from SW480 cells transfected with miR-10a mimics or NC. Experiments were performed at least in triplicate and the results are presented as the mean ± SD. Student's t-test was used to analyze the data. *P<0.05, **P<0.01 vs. NC. NHLFs, normal human lung fibroblasts; miR-10a, microRNA-10a; NC, negative control; OD, optical density.
Discussion
It is well known that cancer metastasis is a very complex process. Various hypotheses have been proposed to explain this biological process. It has been proposed that metastatic dissemination largely depends on mechanical factors that result from the anatomical structure of the vascular system (30). In addition, it has been reported that cancer metastasis is initiated by numerous subpopulations of cells that have different biological characteristics, including metastatic potential (31). The most accepted hypothesis is Paget's ‘seed and soil’ hypothesis, which suggests that the outcome of metastasis is not due to chance, but due to certain tumor cells having specific affinity for the microenvironment of certain organs (32). Previously, the ‘seed and soil’ hypothesis was revised, and it has been proposed that primary neoplasms and metastases consist of both tumor cells and microenvironment cells (33). In the present study, fibroblasts were used, which are dominant, long-lived and highly plastic cells present within the tumor microenvironment. The experiments aimed to determine the mechanism by which primary tumor cell exosomal-miR-10a regulates lung metastasis in patients with CRC.
Previous studies have suggested that miR-10a expression is upregulated in various tumor types, including breast (34), pancreatic cancer (35), hepatocellular carcinoma (36) and non-small cell lung cancer (NSCLC) (37,38). In addition, higher expression levels of miR-10a have been reported to be positively correlated with advanced tumor stage and positive lymph node metastasis in NSCLC (38). In contrast to these observations, a different role of miR-10a was reported in our previous study, which revealed a negative correlation between miR-10a expression and distant metastasis and invasion depth in patients with CRC (21). In the present study, results indicated that miR-10a expression was lower in the serum samples of patients with CRC compared with the levels noted in healthy subjects. In addition, lower expression levels of miR-10a were detected in cancer tissues compared with those noted in the paired adjacent normal tissues of patients with CRC. The expression of miR-10a was also inversely associated with the invasion depth of CRC. The different functions of miR-10a in various types of cancer may be due to the biological heterogeneity of tumor cells. Due to these differences, our future studies aim to provide a deeper understanding of the effects of miR-10a on lung metastasis of CRC. Additional experiments will be conducted in the future to clarify the mechanism underlying lower miR-10a expression in the serum of patients with CRC.
To address the role of miR-10a in the process underlying lung cancer metastasis of CRC, NHLFs were established and identified. Fibroblasts are associated with all stages of disease progression, including cancer metastasis, and they are considered a component of the general host response to tissue damage caused by cancer cells (39). Notably, a more accurate definition of a fibroblast is a resting mesenchymal cell with the potential to be activated by appropriate stimuli and thus become a mesenchymal stem cell (39). In the present study, exosomes were used, which are considered a significant factor within the microenvironment, to investigate the specific function of miR-10a on regulating the crosstalk between the primary CRC tumor site and lung metastatic loci. The results revealed that miR-10a in exosomes secreted by CRC cells could suppress the proliferative activity of NHLFs. In contrast to these findings, miR-10a-overexpressing exosomes promoted cell proliferative activity within 2 days, and there was no significant difference in proliferation between the miR-10a and NC groups after 2 days, which was possibly caused by non-continuous supplementation of exosomes. In addition, the data demonstrated that miR-10a reduced the migratory ability of NHLFs within 48 h, as determined by wound-healing and Transwell assays. These reduced proliferative and migratory capabilities of NHLFs may be adverse factors for CRC primary cell metastasis.
IL-6 is a pro-inflammatory cytokine that has been positively associated with tumor progression and metastasis in various types of cancer (40,41). It has been reported that high levels of IL-6 and IL-8 in the circulation are associated with decreased overall survival of patients with melanoma (42). The present study demonstrated that exosomal-miR-10a derived from SW480 cells reduced IL-6 and IL-8 expression levels in NHLFs. IL-1β facilitates invasion and extravasation in the early stages of metastasis (43), and promotes EMT (44). The IL-1β inflammatory response has been reported to be driven by primary breast cancer and is considered to be dissemination-supportive and colonization-suppressive factor, thus leading to the suppressed metastasis of breast cancer (45). Moreover, microenvironment-secreted IL-1β may promote breast cancer metastatic colonization in the bone via activation of Wnt signaling (46). In the present study, IL-1β expression was downregulated in NHLFs treated with exosomes from SW480 cells overexpressing miR-10a, thus suggesting that the IL-1β expression of NHLFs may mediate an inflammatory response, which may partly contribute to CRC cell metastasis. The present study was limited by the lack of evaluation of downstream pathway mechanisms of miR-10a, which could reduce proliferation and migration of NHLFs and decrease the expression of inflammatory cytokines. The exact mechanism will be assessed in follow-up studies.
In conclusion, the present study provided evidence that miR-10a was expressed at lower levels in the serum and cancer tissues of patients with CRC, and its expression was inversely associated with invasion depth of CRC. Furthermore, exosomal-miR-10a derived from CRC cells reduced the proliferative and migratory activities, and the expression of IL-6, IL-8 and IL-1β, in NHLFs. These data may provide a novel insight into the mechanism underlying lung metastasis and may provide molecular biomarkers for the assessment of tumor metastasis to the lung tissues of patients with CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Talent Training Subsidy Project of Hebei Province (grant no. A201902029), the Natural Science Foundation of Hebei Province (grant no. H2018105049) and the Science and Technology Project of Tangshan City (grant no. 19150246E).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YufengL and YankunL conceived the project and supervised the experiments. JW, YingL and XZ performed the cell experiments and extraction exosomes assay. YuantingL and JG performed the surgery to collect the samples and performed the RT-qPCR assay. YanL, ZW and JZ collected the clinical serum and tissue samples and the information of patients with CRC for the experiments, and performed the FACS assay. YW and WH performed statistical analysis. YankunL and YufengL wrote the manuscript with help from all of the authors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The use of clinical samples was approved by the Ethical Committee of Tangshan People's Hospital, and written informed consent was obtained from the individuals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhu J, Tan Z, Hollis-Hansen K, Zhang Y, Yu C, Li Y. Epidemiological trends in colorectal cancer in China: An ecological study. Dig Dis Sci. 2017;62:235–243. doi: 10.1007/s10620-016-4362-4. [DOI] [PubMed] [Google Scholar]
- 2.Chakedis J, Schmidt CR. Surgical treatment of metastatic colorectal cancer. Surg Oncol Clin N Am. 2018;27:377–399. doi: 10.1016/j.soc.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Engstrand J, Nilsson H, Strömberg C, Jonas E, Freedman J. Colorectal cancer liver metastases-a population-based study on incidence, management and survival. BMC Cancer. 2018;18:78. doi: 10.1186/s12885-017-3925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siebenhüner AR, Güller U, Warschkow R. Population-based SEER analysis of survival in colorectal cancer patients with or without resection of lung and liver metastases. BMC Cancer. 2020;20:246. doi: 10.1186/s12885-020-6710-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Najafi M, Mortezaee K, Majidpoor J. Stromal reprogramming: A target for tumor therapy. Life Sci. 2019;239:117049. doi: 10.1016/j.lfs.2019.117049. [DOI] [PubMed] [Google Scholar]
- 7.Rønnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: Importance of the stromal reaction. Physiol Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Song E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18:99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Ma S, Xiang M, Tong S. HTRA1 promotes transdifferentiation of normal fibroblasts to cancer-associated fibroblasts through activation of the NF-κB/bFGF signaling pathway in gastric cancer. Biochem Biophys Res Commun. 2019;514:933–939. doi: 10.1016/j.bbrc.2019.05.076. [DOI] [PubMed] [Google Scholar]
- 10.Affo S, Yu LX, Schwabe RF. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol. 2017;12:153–186. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javeed N, Mukhopadhyay D. Exosomes and their role in the micro-/macro-environment: A comprehensive review. J Biomed Res. 2017;31:386–394. doi: 10.7555/JBR.30.20150162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S, Lin HY, Chou CY, Huang CH, Small J, Sadik N, Ayinon CM, Lansbury E, Cruz L, Yekula A, et al. Navigating the landscape of tumor extracellular vesicle heterogeneity. Int J Mol Sci. 2019;20:1349. doi: 10.3390/ijms20061349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, Xu YM, Huang LF, Wang XZ. Exosomes: Novel biomarkers for clinical diagnosis. ScientificWorldJournal. 2015;2015:657086. doi: 10.1155/2015/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: Formation-function-future. Int J Mol Sci. 2016;17:2028. doi: 10.3390/ijms17122028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, Li G, Tang J, Xiang J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. 2019;18:40. doi: 10.1186/s12943-019-0959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Zhang Y, Wu H, Li Y, Zhang Y, Liu M, Li X, Tang H. miR-10a suppresses colorectal cancer metastasis by modulating the epithelial-to-mesenchymal transition and anoikis. Cell Death Dis. 2017;8:e2739. doi: 10.1038/cddis.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more ‘personalized’ approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Han S, Li Y, Liu Y, Zhang D, Li Y, Zhang J. MicroRNA-20a contributes to cisplatin-resistance and migration of OVCAR3 ovarian cancer cell line. Oncol Lett. 2017;14:1780–1786. doi: 10.3892/ol.2017.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 26.desJardins-Park HE, Foster DS, Longaker MT. Fibroblasts and wound healing: An update. Regen Med. 2018;13:491–495. doi: 10.2217/rme-2018-0073. [DOI] [PubMed] [Google Scholar]
- 27.Spaw M, Anant S, Thomas SM. Stromal contributions to the carcinogenic process. Mol Carcinog. 2017;56:1199–1213. doi: 10.1002/mc.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strutz F, Okada H, Lo CW, Danoff T, Carone RL, Tomaszewski JE, Neilson EG. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol. 1995;130:393–405. doi: 10.1083/jcb.130.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu SC, Lu HH, Fan HC, Wang HW, Chen HK, Lee FP, Yu CJ, Chu YH. The identification of the TRPM8 channel on primary culture of human nasal epithelial cells and its response to cooling. Medicine (Baltimore) 2017;96:e7640. doi: 10.1097/MD.0000000000007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing J. WB Saunders; Philadelphia, London: 1928. Neoplastic diseases; a treatise on tumors PA. [Google Scholar]
- 31.Fidler IJ. Critical factors in the biology of human cancer metastasis: Twenty-eighth G.H.A. Clowes memorial award lecture. Cancer Res. 1990;50:6130–6138. [PubMed] [Google Scholar]
- 32.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 33.Seretis F, Seretis C, Youssef H, Chapman M. Colorectal cancer: Seed and soil hypothesis revisited. Anticancer Res. 2014;34:2087–2094. [PubMed] [Google Scholar]
- 34.Chang CH, Fan TC, Yu JC, Liao GS, Lin YC, Shih AC, Li WH, Yu AL. The prognostic significance of RUNX2 and miR-10a/10b and their inter-relationship in breast cancer. J Transl Med. 2014;12:257. doi: 10.1186/s12967-014-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, Toma H, Nakamura M, Nagai E, Hashizume M, Tanaka M. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol. 2012;19:2394–2402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
- 36.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 37.Yu T, Liu L, Li J, Yan M, Lin H, Liu Y, Chu D, Tu H, Gu A, Yao M. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget. 2015;6:30239–30250. doi: 10.18632/oncotarget.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao M, Pan S, Yang W, Chen S, Shan Y, Shi H. Serum miR-10a-5p and miR-196a-5p as non-invasive biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2018;11:773–780. [PMC free article] [PubMed] [Google Scholar]
- 39.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 40.Ara T, Declerck YA. Interleukin-6 in bone metastasis and cancer progression. Eur J Cancer. 2010;46:1223–1231. doi: 10.1016/j.ejca.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lou W, Ni Z, Dyer K, Tweardy DJ, Gao AC. Interleukin-6 induces prostate cancer cell growth accompanied by activation of stat3 signaling pathway. Prostate. 2000;42:239–242. doi: 10.1002/(SICI)1097-0045(20000215)42:3<239::AID-PROS10>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 42.Tobin RP, Jordan KR, Kapoor P, Spongberg E, Davis D, Vorwald VM, Couts KL, Gao D, Smith DE, Borgers JSW, et al. IL-6 and IL-8 Are linked with myeloid-derived suppressor cell accumulation and correlate with poor clinical outcomes in melanoma patients. Front Oncol. 2019;9:1223. doi: 10.3389/fonc.2019.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spiegel A, Brooks MW, Houshyar S, Reinhardt F, Ardolino M, Fessler E, Chen MB, Krall JA, DeCock J, Zervantonakis IK, et al. Neutrophils suppress intraluminal NK cell-mediated tumor cell clearance and enhance extravasation of disseminated carcinoma cells. Cancer Discov. 2016;6:630–649. doi: 10.1158/2159-8290.CD-15-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 45.Castaño Z, San Juan BP, Spiegel A, Pant A, DeCristo MJ, Laszewski T, Ubellacker JM, Janssen SR, Dongre A, Reinhardt F, et al. IL-1β inflammatory response driven by primary breast cancer prevents metastasis-initiating cell colonization. Nat Cell Biol. 2018;20:1084–1097. doi: 10.1038/s41556-018-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyre R, Alférez DG, Santiago-Gómez A, Spence K, McConnell JC, Hart C, Simões BM, Lefley D, Tulotta C, Storer J, et al. Microenvironmental IL1β promotes breast cancer metastatic colonisation in the bone via activation of Wnt signalling. Nat Commun. 2019;10:5016. doi: 10.1038/s41467-019-12807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.