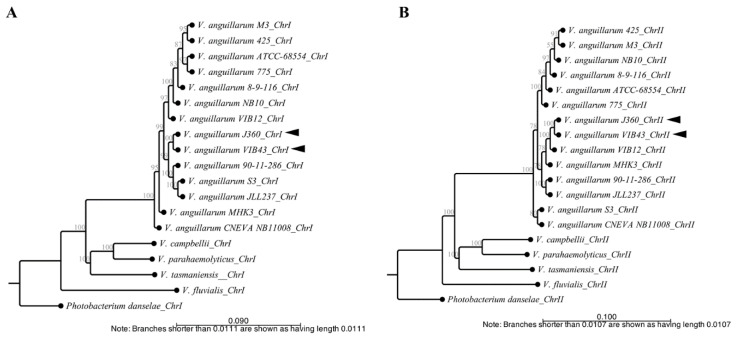
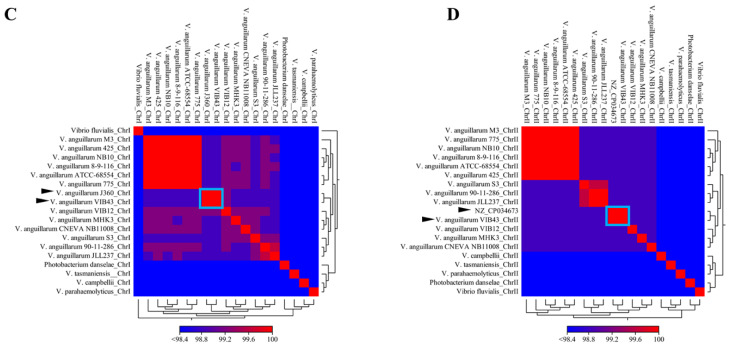
Figure 4.
Phylogenetic and comparative genomic analysis of V. anguillarum J360. (A) Phylogenetic history of V. anguillarum J360 chromosome-I. (B) Phylogenetic history of chromosome-II. Evolutionary history was inferred using the neighbor-joining method, with a bootstrap consensus of 500 replicates for taxa analysis. Evolutionary distance was computed using the Jukes–Cantor method. All ambiguous positions were removed for each sequence pair (pairwise deletion option). (C) Heat map visualization of aligned sequences identity for V. anguillarum J360 chromosome-I. (D) Heat map visualization of aligned sequences identity for V. anguillarum J360 chromosome-II. Whole genome alignments and the phylogenetic analysis involved 18 Vibrio sp. listed in Table 1. Photobacterium damselae 91–197 as an outgroup. Analysis was performed using CLC workbench v.20 (CLC Bio). Black arrows represent V. anguillarum J360 genome and V. anguillarum VIB43 as the closest related strain. Light blue square represents the percentage of identity between strains.