Abstract
Enterococcus faecalis (E. faecalis) is regarded as the major pathogen for persistent periapical periodontitis. The aim of the present study was to investigate the role of antisense walR RNA in the regulation of adjacent downstream genes. Reverse transcription-PCR assays were performed to validate walR. Adjacent downstream genes walK, EF1195, EF1196, and EF1197 were co-transcribed and detect antisense walR RNA. Northern blotting and 5'-rapid amplification of cDNA ends (5'-RACE) assays were conducted to detect and confirm a novel walR antisense (ASwalR) RNA. ASwalR overexpression mutants were constructed, and the biofilm biomass was determined using a crystal violet microtiter assay. The present study detected and confirmed a 550-bp noncoding antisense RNA with the potential to attenuate the activities of the essential response regulator WalR. The levels of antisense walR RNA transcripts were inversely associated with the production of WalR protein. It was showed that overexpression of ASwalR leads to reduced biofilm formation and exopolysaccharide synthesis. Furthermore, the pathogenicity of E. faecalis was markedly decreased by ASwalR overexpression in an in vivo periapical periodontitis model. In summary, the present study detected a novel antisense walR RNA that leads to a reduction in biofilm formation and the pathogenicity of E. faecalis. Collectively, the data suggest a role for ASwalR as a post-transcriptional modulator of the WalR regulator in E. faecalis.
Keywords: WalR, antisense RNA, Enterococcus faecalis, pathogenicity, post-transcriptional control
Introduction
Enterococcus faecalis
(E. faecalis), a generally commensal organism, has emerged as the major pathogen for persistent periapical periodontitis (1). Nonpathogenic (or commensal) bacterial pathogens can switch from the commensal stage to a pathogenic state (2). The virulence of E. faecalis is derived from its ability to sense and adapt to varying environmental stresses during colonization and in the host (3). During long-term interaction with the host, the two-component signal transduction system (TCS) of E. faecalis regulates the expression of virulence genes in response to microenvironmental conditions, including the host environment, and binds to the regulatory regions of target genes accordingly (4). These two-component signal transduction systems are involved in various cellular processes, including cell viability, virulence, biofilm formation, quorum sensing and antibiotic resistance (5). A typical TCS consists of a histidine sensory kinase membrane receptor and its cognate response regulator (6). The histidine kinase is activated by recognition of environmental stimuli, including pH, oxidative stress, antibiotic pressure and nutrient starvation, and relays the activated phosphoryl group to the response regulator for gene expression modulation by binding at the regulatory regions of target genes (4).
The WalRK (also known as VicRK or YycFG) TCS originally from Bacillus subtilis is highly conserved in Gram-positive bacteria and is annotated as VicRK or YycFG in some genera, such as Staphylococcus aureus, Enterococcus faecalis, and Streptococcus mutans (S. mutans) (6). The system WalRK has been proposed because of its major roles in the regulation of genes associated with cell wall synthesis and biofilm formation (7). The essential gene walR, is closely associated with bacterial growth, virulence and biofilm extracellular polysaccharide synthesis and aggregation (1). Bacterial growth and biofilm aggregation are strongly associated with the process of human infections (4). Consistently, the ability of E. faecalis to form the three-dimensional biofilm scaffold contributes to the failure of persistent infected root canal treatments (8). In E. faecalis, it was reported that the inability to construct walR deletion mutants indicated that this regulatory gene was essential for bacterial viability (4).
A noncoding antisense RNA (AS RNA) can bind to the target messenger RNA (mRNA) by base-pairing, and their interaction forms an RNA duplex structure (9). This AS RNA-induced duplex complex generally inhibits mRNA transcription or translation and performs regulatory functions (10). Using high-throughput transcriptomics analyses, AS RNA regulators can be identified in bacteria (11). An endogenous vicR antisense RNA transcript has been identified in S. mutans (12). In addition, it was found that the production of VicR protein associated inversely with different levels of vicR antisense RNA and that the biofilm biomass decreased in the vicR antisense overexpression (12). Considering the close proximity of E. faecalis and S. mutants in related Gram-positive cocci, it was suggested that there were similar structural and functional relationships between vicR in S. mutans and walR in E. faecalis. Additionally, by mapping the E. faecalis walR gene with BLAST searches, a high similarity (the DNA sequences are 70% identical) with the homologous vicR gene in S. mutans was demonstrated. In the present study, a potential ASwalR was hypothesized, and whether the potential ASwalR was specifically associated with regulation of WalR function was investigated in E. faecalis.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in the present study are listed in Table SI. E. faecalis strains purchased from Guangdong Huankai Microbial Science & Technology Co., Ltd. were grown in brain heart infusion (BHI) broth (BD Biosciences) at 37˚C in a 5% CO2 atmosphere with 500 µg/ml spectinomycin when ASwalR overexpression strains were incubated at 37˚C overnight. E. faecalis strains were cultured to mid-exponential phase (optical density at a wavelength of 600 nm of OD value=0.5). For antisense overexpression strain construction, the shuttle vector pDL278 (Novagen) was used to overexpress ASwalR or walR sequence under the control of the walR gene promoter region. First, the antisense walR or walR sequences were obtained by oligonucleotide synthesis (Sangon Biotech Co., Ltd.). It was reported that E. faecalis V583 carrying the empty vector pDL278 has no effects of the vector itself on the phenotypes examined (13). Next, antisense ASwalR or walR sequences were cloned into the pDL278 plasmid at BamHI and EcoRI restriction sites. All recombinant ASwalR RNA or walR gene overexpression plasmids were verified by PCR and DNA sequencing analyses. The recombinant plasmid pDL278ASwalR or pDL278walR was transformed into E. faecalis V583 strains, as previously described (1,12).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNAs were purified from E. faecalis strains using a MasterPure RNA purification kit (Epicentre; Illumina Inc.). Residual genomic DNA was removed using Turbo RNase-free DNase I (Ambion; Thermo Fisher Scientific, Inc.). The quality and integrity of RNAs were assessed by 1% agarose gel electrophoresis. The concentration and purity of RNAs were determined using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Inc.). Total RNAs were extracted from E. faecalis V583, ASwalR and walR overexpression strains as aforementioned. Subsequently, purified RNA was reverse transcribed to cDNA using a PrimeScript RT Reagent kit (Takara Bio, Inc). RT-PCR was conducted for identification and detection of antisense RNA. For ASwalR detection, a gene-specific primer (PCR1) was used for the first strand cDNA synthesis, and another gene-specific primer (AS2) was used for RT-qPCR analysis (Supplementary Table II). The expression of walR and walK and virulent factor genes including ace (adhesin of collagen), esp (protein surface), epal, epaA (enterococcal polysaccharide antigen), and gel (gelatinase) of all E. faecalis strains were also examined. RT-qPCR was performed using primers listed in Supplementary Table II on a LightCycler 480 (Roche Diagnostics), following the instructions of the PrimeScript RT Reagent kit (Takara Bio, Inc.). RT-qPCR conditions were: 95°C for 30s, followed by 40 cycles at 95°C for 5s; 60°C for 30s, and then dissociation. The threshold cycle values (CT) were quantified and transcription levels of each gene were compared to the expression of the 16sR gene, which was used as a reference gene (1).
Reverse transcription-PCR (RT-PCR) assays for walR co-transcribed operator and antisense RNA detection
Conditions for PCR reactions were: 94˚C, 3 min (initial denaturation); 32 cycles at 94˚C, 30 sec (denaturation); 55˚C, 40 sec (primer annealing); and 72˚C, 1 min (primer extension). For potential antisense walR RNA detection, total RNAs were purified from E. faecalis strains after planktonic growth using a MasterPure RNA purification kit (Epicentre; Illumina Inc.). Residual genomic DNA was removed using Turbo RNase-free DNase I (Ambion; Thermo Fisher Scientific, Inc.). The quality and integrity of RNAs were assessed by 1% agarose gel electrophoresis. The concentration and purity of RNAs were determined using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Inc.).
Total RNA was prepared from strain E. faecalis V583 grown as planktonic culture in BHI and used as templates to validate walR and adjacent downstream genes, walK, EF1195, EF1196, and EF1197 were co-transcribed (12). The primers for PCR amplification were presented in Table SIII. For first-strand DNA synthesis, walR antisense-(PCREf) and sense-specific primers (ASEf) were used. The ASwalR RT-PCR product was purified with QIAquick PCR Purification Kit according to the manufacturer's instructions and subsequently sequenced. Detection of antisense RNA was performed by 1% agarose gel electrophoresis and RT-PCR assays were performed using a first strand-specific primer for synthesis of cDNA transcripts using the First Strand cDNA synthesis kit (Thermo Fisher Scientific, Inc.) (14). The primers (PCREf) used for first strand synthesis PCR are listed in Table SII. Following the reverse transcription reaction, the first-stand cDNA synthesis products were used as templates. The sense strand primer (ASEf) for PCR amplification is shown in Table SIV.
Northern blotting
Northern blotting assays were performed as previously described with minor modifications (12,15). In brief, 10 µg of total RNA was purified from E. faecalis planktonic cultures. The RNA samples were detected by electrophoresis in 1% formaldehyde-agarose gels and transferred to nylon membranes (Amersham; Cytiva). The probe was made specifically (listed in Table SIV) and labeled using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics). Blots were incubated 16 h with 20 ng/ml probes at 50˚C, and signals were detected using the CSPD Star substrate (Roche Diagnostics).
5'-rapid amplification of cDNA ends (5'-RACE) assay
To find the transcription initiation site and probable termination site of ASwalR, total RNA (20 µg) from E. faecalis strains was ligated to the 5'-RACE outer adapter from the FirstChoice RLM-RACE Kit (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions (12). The primers used for PCR assays for 5'-RLM-RACE included 5'-RACE gene-specific outer primer and 5'-RACE gene-specific inner primer as listed in Table SIV. The nested PCR thermocycling conditions were as follows: Initial activation of 95˚C for 3 min; followed by 35 cycles of denaturation at 94˚C for 30 sec, annealing at 56˚C for 30 sec and extension at 72˚C for 1 min; and a final extension at 72˚C for 7 min was performed thereafter. An aliquot (6 µl) of the PCR mixture was assessed by 1% agarose gel electrophoresis using reactants without a cDNA template as the control and sequencing by Sangon Biotech Co., Ltd..
Western blotting
Protein extraction and western blotting were proceeded as previously described (12,13). E. faecalis cells grown as planktonic cultures of mid-exponential phase were washed and resuspended in 10 mM Tris-HCl buffer (pH 8.0). Cells were mechanically disrupted by ultrasonication with glass beads (diameter, 0.1 mm) for three cycles of 15 sec with 1 min rest on ice. Clear supernatants were collected by centrifugation (12,000 x g, 2 min, 4˚C) and protein concentrations were determined using a Bradford assay (BioRad Laboratories, Inc.). For western blotting, equal amounts of protein (30 µg) were mixed with Laemmli sample buffer (BioRad Laboratories, Inc.) in boiling water for 10 min. The protein samples were loaded on precast 4-20%, gradient gels (Beijing Solarbio Science & Technology Co., Ltd.) and electrotransferred to PVDF membranes (Thermo Fisher Scientific, Inc.). Polyclonal antibodies against r-WalR were produced using the standard 70 days Rabbit Protocol (AbMax Biotechnology Co., Ltd.). The membranes were blocked, then probed with purified WalR-specific rabbit antibody (AbMax Biotechnology Co., Ltd.; http://www.antibodychina.com/, cat. no. scu001; 1:1,000) for 2 h at room temperature and incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (HRP conjugated, cat. no. SSA004; 1:10,000) for 2 h at room temperature. A BioRad GS-700 Imaging Densitometer (BioRad Laboratories, Inc.) was used to determine the signal density of protein signals.
Biofilm assessment
E. faecalis V583, ASwalR and walR+ strains were cultured in BHI for 24 h at 37˚C as previously described (1,12,13). Crystal violet assay was performed to measure the biomass of each E. faecalis biofilm. E. faecalis V583 biofilms were set as controls. Briefly, the biofilms were washed three times with PBS. Subsequently, the biofilms were stained with 0.1% (w/v) crystal violet for 15 min at 37˚C and destained with 1 ml ethanol/acetone (at a ratio of 8:2) solution. The collected solution was removed to a new plate and measured with a microplate reader (ELX800; Gene Company Ltd.) at a wavelength of 600 nm. For confocal laser scanning microscopy (CLSM), the E. faecalis stains in biofilm were labeled with SYTO9 (Invitrogen; Thermo Fisher Scientific, Inc.) and the extracellular polymeric substance (EPS) matrix was stained with an Alexa Fluor 647-labeled dextran conjugate (Invitrogen; Thermo Fisher Scientific, Inc.) (1). Three-dimensional reconstruction of the biofilms and the EPS/bacterial ratio were analyzed using Imaris 7.0 software (Bitplane; Oxford Instruments).
Periapical periodontitis lesions in animal experiments
Animal experiments were approved by the Biomedical Research Ethics Committee of West China Hospital (approval no. 2019128A) and conducted according to the guidelines of Institutional Animal Care and Use Committee (IACUC) for animal care and use of laboratory animals (16). A total of 20 6-week-old female Sprague-Dawley rats (260-280 g) were included in the study and anesthetized with ketamine/xylazine (90 and 10 mg/kg, respectively) by intraperitoneal injection. On the occlusal surface of the left mandible first molars, an access opening was made using a high-speed round bur (Aseptico Inc.). The right first mandible molar was used as a blank control. Next, the root canal preparation was performed with sterile K-files (0.02 taper, sizes #15-#35). Subsequently, 50 µl log-phased E. faecalis V583 bacterial suspensions were inoculated into the pulp chamber and sealed with light-cured flowable resin (3M Filtek™ Z350XT; 3M) in the V583 group (n=10). For the ASwalR group (n=10), 50 µl log-phased ASwalR E. faecalis bacterial suspensions were applied for establishment of an infection model. After 4 weeks of operation, the rats were scarified by euthanasia under deep anesthesia using ketamine/xylazine by cervical dislocation, and the rats were imaged using the Quantum GX Micro-CT System (PerkinElmer, Inc.). The scanning conditions were as follows: kV=90; CT µA=72; 360˚ scan time=8 sec. The reconstructed images were analyzed with Analyze 12.0 (PerkinElmer, Inc.).
HE and Gram staining and PNA-FISH
For histological evaluation and peptide nucleic acid fluorescence in situ hybridization (PNA-FISH), the mandibular bone was longitudinally split into two parts. The specimens were prepared for histological evaluation. The mandibular bone was fixed in 10% neutral buffered formalin for 72 h at room temperature, decalcified in 10% EDTA and embedded in paraffin. The 5-µm slices were prepared for hematoxylin and eosin (HE) and Gram staining for tissue assessment with a light microscope at x40 magnification. For PNA-FISH, bone specimens were also fixed for 72 h in 10% neutral buffered formalin and decalcified in 10% EDTA decalcified for 3 weeks before being embedded in paraffin blocks and sectioned to 3 µm thickness. Smears were deparaffinized in xylene (2x5 mins) and rehydrated by a graded ethanol series (100, 95, 80, 70 and 50%) for 5 min each time and permeated with proteinase K (Wuhan ServiceBio Technology Co., Ltd.) at 37˚C for 25 min. Finally, all smears were washed up with distilled water for 10 min and allowed to air dry.
The above slides were fixed in 100% methanol for 10 min at 37˚C, followed by immersion in 80% ethanol for 10 min. For pre-hybridization, one drop of hybridization solution without probe containing 10% (w/v) dextran sulfate (Thermo Fisher Scientific, Inc.), 10 mM NaCl (Panreac), 0.2% (w/v) polyvinylpyrrolidone (Sigma-Adrich; Merck KGaA), 0.2% (w/v) Ficoll (Thermo Fisher Scientific, Inc), 5 mM disodium EDTA (PanReac AppliChem), 0.1% (v/v) Triton X-100 (PanReac AppliChem), 50 mM Tris-HCl (pH 7.5; Thermo Fisher Scientific, Inc.) was applied for 1 h at 37˚C. From the template of genomic DNA a fluorescein amidite-labeled PNA probe (5'-GGTGTTGTTAGCATTTCG-3') targeting E. faecalis 16S rRNA (Wuhan Servicebio Technology Co., Ltd.) was applied. The slides were then incubated with hybridization solution containing 200 nM PNA probe overnight at 37˚C. Subsequently, the slides were co-cultured in saline sodium citrate washing buffer (Wuhan ServiceBio Technology Co., Ltd.) at 37˚C for 30 min and dried in air. Bone smears were then stained with 100 µl DAPI (Thermo Fisher Scientific, Inc.) in the dark for 30 min at room temperature. The samples were washed with PBS buffer twice and dried in air. The slides were mounted with Gold Antifade reagent (Thermo Fisher Scientific, Inc.) and stored at 4˚C for further investigation with fluorescent microscope at 40 magnification.
Statistical analysis
Bartlett's test was conducted to analyze the homogeneity of data variances and Shapiro-Wilk test was applied to determine the normal distribution of data. For parametric testing, one-way ANOVA was performed followed by pairwise multiple comparisons of Tukey's test using SPSS software 18.0 (SPSS, Inc.). Data are presented as the mean ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
WalR gene operon contains an antisense walR RNA
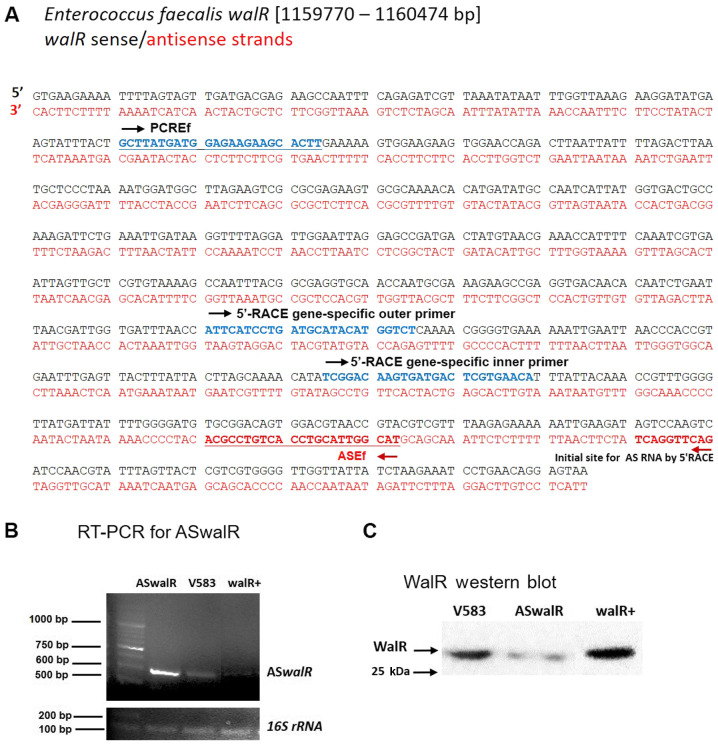
Downstream of the walR (EF1193) in E. faecalis V583, a cluster of four genes (EF1194 to EF1197) is co-transcribed (Fig. 1A). Using a series of overlapping primers, RT-PCR indicated co-transcription comprising walR and the four downstream genes (Fig. 1B), indicating that walR, walK, EF1195, EF1196, and EF1197 are part of the WalR regulon. RNA agarose gel electrophoresis for northern blotting was included in Fig. S1.
Figure 1.
WalR regulon and downstream genes in Enterococcus faecalis V583. (A) Schematic of the gene locus and RT-PCR primer sets. Downstream of walR, a cluster of five genes (EF1194 to EF1197) is similarly transcribed in the same orientation. (B) RT-PCR with lanes numbered 1-4 according to primer sets used. Total RNA was used as the negative control for all the PCR reactions, indicating RT was not contaminated with genomic DNA. RT, reverse transcription; F, forward; R, reverse. RNA agarose gel electrophoresis for Northern blotting was included in the Supplementary Fig. 1.
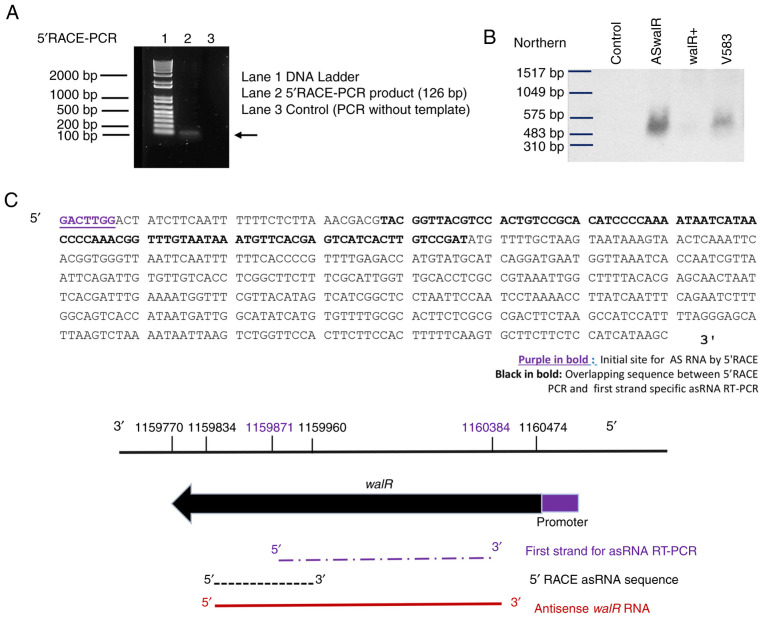
To investigate whether a potential antisense RNA was specifically associated with walR, first strand cDNA synthesis was performed to detect the transcript in E. faecalis V583. The sense and antisense sequences of the walR gene (EF1193) are shown in Fig. 2A. To determine the 5'-terminus of ASwalR RNA, 5'-RACE assays of ASwalR were conducted. The primers used and the design and position of primers for 5'-RACE is indicated in Fig. 2A. The sequencing analyses indicated that the potential ASwalR contained 550 bp of antisense sequence in the E. faecalis V583 genome by RT-PCR assays (Fig. 2B). Western blotting showed that the expression of WalR protein was inversely associated with different levels of ASwalR transcripts (Fig. 2C). The size of the 5'-RACE PCR product was ~120 bp (Fig. 3A). Additionally, the differences in ASwalR abundance in E. faecalis planktonic growth were confirmed by northern blotting (Fig. 3B). The sequence predicted that the 5'-terminus of ASwalR begins at and covers most of the walR coding sequence (Fig. 3C).
Figure 2.
Detection of walR antisense RNA. (A) Primer design for detection of antisense walR RNA. (B) Differences in ASwalR RNA abundance in Enterococcus faecalis cultures by first cDNA strand synthesis and RT-PCR using walR gene-specific anti-sense and sense strand primers. The 16sR gene was used as an internal standard. (C) WalR production blot was quantified in the cells grown as aforementioned from western blots probed with anti-WalR antibody. PCREf, walR antisense-specific primer; ASEf, walR sense-specific primer; 5'-RACE, 5'-rapid amplification of cDNA ends; AS, antisense; RT-PCR, reverse transcription-PCR; ASwalR, walR antisense RNA.
Figure 3.
Structure of the ASwalR transcript and association with walR mRNA. (A) Detection of the 5'-terminus of the ASwalR transcript by 5'-RACE. The PCR product size was ~120 bp. (B) Transcript amounts of ASwalR in Enterococcus faecalis cultures were observed in a representative northern blot. The loading sample without RNA was used as the blank control. (C) Schematic of the ASwalR 5'-terminus showing that transcription starts within the walR ORF and is complementary to most of the walR ORF. The sequence of the first strand synthesis ASwalR RT-PCR amplicon was from 1159871-1160384 bp. The overlapping sequence from first strand-specific ASwalR RT-PCR and 5'-RACE product (89 bp) was from 1159871-1159960 bp in the genome. The full length for ASwalR RNA is 550 bp (1159834-1160384 bp). ASwalR, walR antisense RNA; ORF, open reading frame; RT-PCR, reverse transcription-PCR; 5'-RACE, 5'-rapid amplification of cDNA ends; AS, antisense.
Antisense walR RNA negatively affects the production of WalR and suppresses biofilm formation
Using CLSM visualization of double-stained biofilms, it was found that overproducing ASwalR markedly decreased EPS production compared with E. faecalis V583 strain, while biofilm cells were packed within the enriched EPS matrix, which was stained red in the walR+ strains, similar to the results obtained with the V583 parent strain (Fig. 4A). These findings were further confirmed by quantitation of data revealing that ASwalR exhibited the lowest EPS/bacterial biomass volume ratio, indicating a role for the walR gene in EPS architecture development (Fig. 4B). Next, the effects of ASwalR overexpression on the ability to form biofilms were evaluated. E. faecalis V583, ASwalR and walR+ strains were allowed to form biofilms in BHI for 24 h. The biomass was quantified by a crystal violet microtiter assay, and the results indicated that overexpression of ASwalR resulted in a 45% decrease in biofilm growth compared with the V583 parent strain (Fig. 4C). However, the biomass of the walR+ strain was slightly but non-significantly increased compared with the E. faecalis V583 biofilm (Fig. 4C). The expression levels of ASwalR and walR were quantified, which indicated that the transcripts of ASwalR in the ASwalR strain were 2.7-fold higher compared with V583 parent cells, while the transformation of a pDL278 empty vector did not affect the levels of walR mRNA (1). The results showed that the levels of transcripts of two glycosyltransferase genes, epal and epaA, were significantly decreased in the ASwalR strain when compared with E. faecalis V583 strain (Fig. 4D).
Figure 4.
ASwalR overexpression suppresses E. faecalis biofilm aggregation and EPS production. (A) Double labeling of the biofilms in the E. faecalis V583, ASwalR and walR+ strains. Green, bacterial cells (SYTO9); red, EPS matrix (propidium iodide, PI). Scale bar, 100 µm. (B) Percentage of EPS matrix/bacteria ratio in biofilm growth. n=10. *P<0.05. (C) Biomass was quantified by crystal violet staining. n=10. *P<0.05. (D) Reverse transcription-PCR analysis showed the gene transcripts in E. faecalis V583, ASwalR, and walR+ strains. E. faecalis gene expression was quantified using 16sR as an internal control and calculated based on the E. faecalis V583 expression, which was set as 1.0. n=10. *P<0.05 when compared to the E. faecalis V583. ASwalR, walR antisense RNA; EPS, exopolysaccharide; E. faecalis, Enterococcus faecalis; OD, optical density; Ace, adhesin of collagen; esp, protein surface; epal, epaA, enterococcal polysaccharide antigen; gel, gelatinase.
WalR antisense inhibited the pathogenicity and reduced Periapical Lesion Size
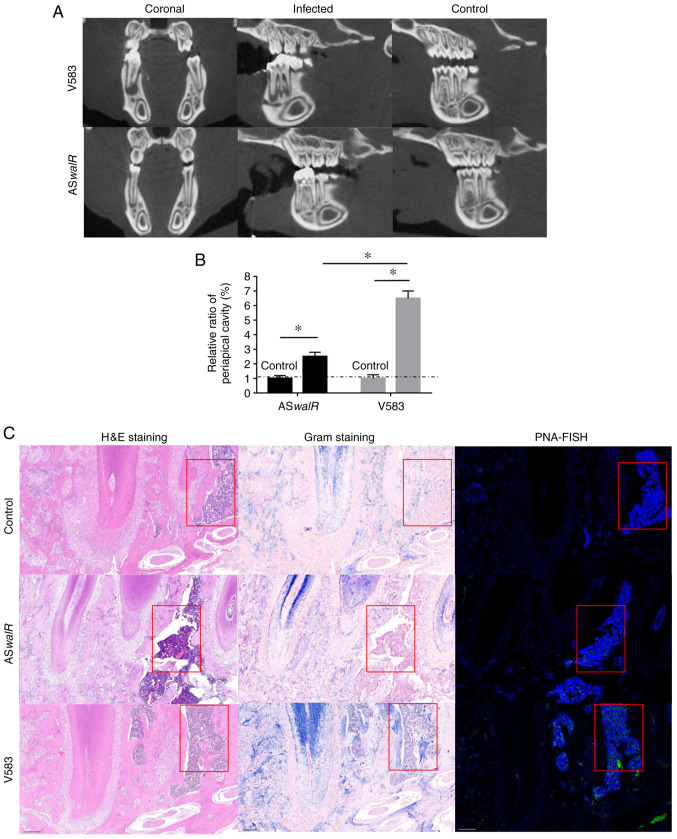
The micro-CT results showed that the severity of bone destruction was markedly higher in the E. faecalis V583-infected group compared with the ASwalR-treated group (Fig. 5A). Quantitatively, the average relative ratio of the periapical cavity was ~2.5% in the ASwalR-treated group compared with the right first mandible molar as a blank control, which was significantly lower than that of E. faecalis V583-treated group (Fig. 5B). This trend indicated that ASwalR-transformed stains presented a limited capability to infarct infected bone tissues. Histological assessments in HE-stained samples showed bone tissue absorption and inflammatory infiltration in the E-faecalis V583-colonized groups, which were more severe compared with control and ASwalR groups (Fig. 5C; left column). Gram staining results showed that more Gram-positive cells were identified within the bone of the E-faecalis V583 group compared with the ASwalR group (Fig. 5C; middle column). After labeling with PNA fluorescent probes, bacterial cells were specifically detected by green fluorescence. The control group indicated no Gram-positive cells were inoculated (Fig. 5C; right column). The fluorescence intensity of the E. faecalis group was higher compared with ASwalR strain-infected samples. The control group indicated no fluorescence intensity of the E. faecalis were observed.
Figure 5.
ASwalR overexpression reduces the periapical lesion size and inhibits bacterial aggregation. (A) Reconstructed images for micro-CT scanning of rat periapical lesions. (B) Relative periapical cavity levels (%) were calculated n=10. *P<0.05. (C) Samples were obtained from infected periapical lesions at 4 weeks. HE-stained histological slices (left lane; scale bars, 100 µm) and Gram-stained samples (middle lane; scale bars, 100 µm). The red boxes indicated inflammatory cells in the HE-staining or bacteria in the Gram-staining. The presence of fluorescent Enterococcus faecalis was identified with a PNA-FISH probe for bacterial 16S rRNA (right lane; scale bars, 100 µm). ASwalR, walR antisense RNA; HE, hematoxylin-eosin; PNA-FISH, peptide nucleic acid- fluorescence in situ hybridization.
Discussion
The WalRK (also known as VicRK or YycFG) two-componen t signal transduction system is highly conserved in Gram-positive bacteria, which originated from Bacillus subtilis (6). In E. faecalis, the downregulation of walR led to a reduction in the dextran-dependent aggregation in the biofilm and inhibited the transcription of virulence genes (1). Several genes in the region of the walR locus were assigned to the WalR operon as RT-PCR results indicated co-transcription comprising walR and the four downstream genes, including walK, EF1195, EF1196, and EF1197. The proximity of walK to walR suggested a close interaction and role in the regulation of WalRK TCS (17). The function annotated for the EF1195 gene, a hypothetical protein belonging to the conserved protein domain family YycH, is associated with the regulatory protein YycH in the two-component signal transduction system YycFG (18). The most distal genes, EF1196 and EF1197, in the extended WalR regulon encoded a putative Yycl protein and a metallo-beta-lactamase, YycJ, which probably plays a role in regulating protein function by lactamization (19). Although the information of these genes regulated by WalR remains limited, the close proximity of the other genes in the extended WalR regulon suggested a functional relationship, including cell wall biogenesis and biofilm formation, which also needs further investigation.
The evidence has revealed that substantial antisense transcripts, including intergenic and intragenic transcription, exist in the bacteria (11). A cis antisense noncoding RNA to the walR gene (ASwalR) was identified and confirmed by northern blotting and 5'-RACE. The effect of ASwalR on decreasing and inhibiting walR gene expression and associated production of WalR was further detected using following RT-PCR and western blotting. Similarly, it was shown that antisense RNA could combine with its reverse complementary sequences and restricted the expression of targeted mRNA (12). Based on this, the present study hypothesized an inverse association between ASwalR levels and walR transcripts and WalR production. The inverse association observed is consistent with the ASwalR-walR mRNA complex blocking its transcription and translation (20).
To determine whether ASwalR RNA activity played a role in biofilm formation, the present study constructed ASwalR and walR overexpression mutant strains to inhibit ASwalR levels. Our previous results suggested that the interference of the walR gene suppressed the expression of virulent genes associated with cellular adhesion and biofilm growth (1). In E. faecalis, glycosyltransferase is a type of enzyme involved in the synthesis of polysaccharides by the transfer of a sugar precursor onto sugar residues (21). Biofilm is one of major factors for infection development and processes (1,13). By overexpression of ASwalR, the levels of two glycosyltransferase genes, epal and epaA were significantly reduced, which contributed to impaired EPS production in the ASwalR biofilm and decreased biomass of ASwalR overproducing strains. In addition, the transcripts of virulence genes were lower in all ASwalR-treated groups. Therefore, the ability of ASwalR strains to induce periapical infections was reduced compared to the parent V583 strains, demonstrated by histological evidence from HE and Gram staining (1). The effects of walR overexpression on both bacterial growth and EPS production were similar to the V583 parental strain levels, justifying additional studies.
These analyses of biofilm formation again confirm that the ASwalR RNA functions in biofilm formation by E. faecalis. Next, the role of ASwalR RNA in the pathogenicity of E. faecalis was validated using a Sprague-Dawley rat model. The results indicated that ASwalR overexpression markedly reduced the periapical lesion size at 4 weeks compared to the E. faecalis V583 group, which presented large periapical lesions. In the present study, restricting the periapical lesions in ASwalR-treated group was considered an important step for stimulating the periapical repair process (22). Additionally, histological evidence from HE and Gram staining demonstrated that the ability of ASwalR strains to cause periapical infections was reduced compared with the parent V583 strains. Furthermore, the presence of E. faecalis bacterial cells involved in periapical tissues was identified by PNA-FISH, which showed higher fluorescence intensity in the parent V583 strains. It was hypothesized that the pathogenicity of E. faecalis was markedly decreased by ASwalR overexpression in periapical periodontitis. However, one limitation of the present study is the lack of direct evidence showing whether ASwalR-mRNA duplexes can be disrupted by additional RNase activities. Bioinformatic studies for the ASwalR transcript started in the opposite direction to walR, which was reverse complementary to the walR transcript (BPROM at http//:Linux1.softberry.com/berry.phtml). A putative-35 box, TTCACA, and a-10 box, TTCTAATCT. Future research is needed to construct a vector which contains the entire ASwalR gene and the promoter of the ASwalR to identify the transcriptional initial site of ASwalR.
In summary, the downstream genes walK, EF1195, EF1196, and EF1197 together with the walR gene are co-transcribed in E. faecalis V583. The present study detected and confirmed a 550-bp noncoding antisense RNA with the potential to attenuate the activities of the essential response regulator WalR. The levels of antisense walR RNA transcripts were inversely associated with the production of WalR protein. In addition, the present study showed that overexpression of ASwalR led to a reduction in biofilm formation and EPS synthesis. Furthermore, the results of animal experiments revealed that the pathogenicity of E. faecalis was markedly decreased by ASwalR overexpression in periapical periodontitis. Collectively, the data suggested a role for ASwalR as a post-transcriptional modulator of the WalR regulator in E. faecalis and revealed that preserving the novel antisense walR RNA will be a potential substitutive therapy for E. faecalis infections, especially in the treatment of periapical periodontitis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science Foundation of China (grant no. 81800964), the Sichuan Provincial Natural Science Foundation of China (grant nos. 2018SZ0125 and 2019YFS0270).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SW contributed to conception, design and interpretation of data, drafted and critically revised the manuscript. YL contributed to design, and data interpretation, drafting manuscript and critical revision the manuscript. LL contributed to conception, design, and interpretation of data, drafted and critically revised the manuscript. HZ contributed to conception, design, and interpretation of data, drafted and critically revised the manuscript. All authors gave their final approval and agree to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The West China Hospital of Sichuan University Biomedical Research Ethics Committee approved the animal experiments for this investigation (approval no. 2019128A).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wu S, Liu Y, Zhang H, Lei L. The susceptibility to calcium hydroxide modulated by the essential walR gene reveals the role for Enterococcus faecalis biofilm aggregation. J Endod. 2019;45:295–301.e2. doi: 10.1016/j.joen.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colomer-Winter C, Flores-Mireles AL, Baker SP, Frank KL, Lynch AJL, Hultgren SJ, Kitten T, Lemos JA. Manganese acquisition is essential for virulence of Enterococcus faecalis. PLoS Pathog. 2018;14(e1007102) doi: 10.1371/journal.ppat.1007102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock LE, Perego M. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J Bacteriol. 2004;186:7951–7958. doi: 10.1128/JB.186.23.7951-7958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol. 2010;13:232–239. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology. 2000;146:1573–1583. doi: 10.1099/00221287-146-7-1573. [DOI] [PubMed] [Google Scholar]
- 7.Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: Cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 8.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 9.Krinke L, Wulff DL. RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA. Genes Dev. 1990;4:2223–2233. doi: 10.1101/gad.4.12a.2223. [DOI] [PubMed] [Google Scholar]
- 10.Saberi F, Kamali M, Najafi A, Yazdanparast A, Moghaddam MM. Natural antisense RNAs as mRNA regulatory elements in bacteria: A review on function and applications. Cell Mol Biol Lett. 2016;21(6) doi: 10.1186/s11658-016-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei L, Stipp RN, Chen T, Wu SZ, Hu T, Duncan MJ. Activity of Streptococcus mutans VicR is modulated by antisense RNA. J Dent Res. 2018;97:1477–1484. doi: 10.1177/0022034518781765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho EC, Donaldson ME, Saville BJ. Detection of antisense RNA transcripts by strand-specific RT-PCR. Methods Mol Biol. 2010;630:125–138. doi: 10.1007/978-1-60761-629-0_9. [DOI] [PubMed] [Google Scholar]
- 14.Stipp RN, Boisvert H, Smith DJ, Höfling JF, Duncan MJ, Mattos-Graner RO. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS One. 2013;8(e58271) doi: 10.1371/journal.pone.0058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S, Huang F, Zhang H, Lei L. Staphylococcus aureus biofilm organization modulated by YycFG two-component regulatory pathway. J Orthop Surg Res. 2019;14(10) doi: 10.1186/s13018-018-1055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couto M, Cates C. Laboratory guidelines for animal care. Methods Mol Biol. 2019;1920:407–430. doi: 10.1007/978-1-4939-9009-2_25. [DOI] [PubMed] [Google Scholar]
- 17.Aakra A, Vebø H, Snipen L, Hirt H, Aastveit A, Kapur V, Dunny G, Murray BE, Nes IF. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob Agents Chemother. 2005;49:2246–2259. doi: 10.1128/AAC.49.6.2246-2259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 19.Wecker P, Klockow C, Ellrott A, Quast C, Langhammer P, Harder J, Glöckner FO. Transcriptional response of the model planctomycete Rhodopirellula baltica SH1(T) to changing environmental conditions. BMC Genomics. 2009;10(410) doi: 10.1186/1471-2164-10-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krinke L, Wulff DL. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987;1:1005–1013. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- 21.Dale JL, Cagnazzo J, Phan CQ, Barnes AM, Dunny GM. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob Agents Chemother. 2015;59:4094–4105. doi: 10.1128/AAC.00344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddiqui YD, Omori K, Ito T, Yamashiro K, Nakamura S, Okamoto K, Ono M, Yamamoto T, Van Dyke TE, Takashiba S. Resolvin D2 induces resolution of periapical inflammation and promotes healing of periapical lesions in rat periapical periodontitis. Front Immunol. 2019;10(307) doi: 10.3389/fimmu.2019.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.