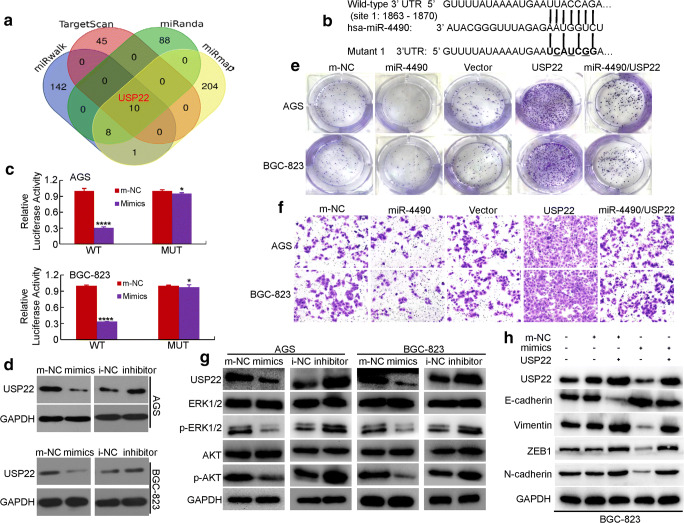
Fig. 4.
USP22 is a direct downstream target of miR-4490 in GC cells. (a) miRmap, miRWalk, miRanda and TargetScan databases were used to predict miR-4490 targets; USP22 was identified as a potential target. (b) Putative miR-4490-binding sequence within the 3′UTR of USP22. Mutations in the complementary site for the seed region of miR-4490 in the 3′UTR of USP22 are indicated. (c) Reporter plasmids containing either the wild-type 3′UTR or a mutated 3′UTR of the USP22 gene were co-transfected into GC cells with a miR-4490-encoding plasmid, after which luciferase activity was measured. WT, wild type; MUT, mutated. *, p > 0.05; ****, p < 0.001. The data are presented as the means ± SD of three experiments. (d) USP22 expression in m-NC and miR-4490 mimic-transfected or i-NC and miR-4490 inhibitor-treated GC cells analyzed by Western blotting. GAPDH was used as a loading control. (e) Representative micrographs of crystal violet-stained cell colonies formed by the indicated GC cells at 12 days after seeding. (f) Assessment of the invasive activity of GC cells transfected with a USP22 expression plasmid and/or miR-4490 mimics. (g) USP22, ERK1/2, p-ERK1/2, Akt, and p-Akt protein expression in GC cells transfected with miR-4490 mimic, inhibitor or NC plasmid. (h) Expression levels of USP22, E-cadherin and vimentin detected by Western blotting in GC cells transfected with USP22 expression plasmid, miR-4490 mimic and/or its control plasmid. The figures are representative of three independent experiments