Abstract
A coronavirus disease (COVID-19) genome surveillance has been conducted at four international airports in Japan, revealing a potential imported COVID-19 risk from multiple countries. The quarantine surveillance based on genome sequencing can enhance sequencing efforts worldwide, as returning travelers may serve as excellent sentinels for the global pandemic.
Keywords: SARS-CoV-2, sentinel surveillance, airports, air travel, genomic, sequencing
In Japan, there has been a sharp increase in the number of coronavirus disease (COVID-19) cases detected among travelers and returnees at airport quarantine stations, despite the pandemic having subsided in parts of many countries. A nasopharyngeal specimen was collected from travelers and returnees arriving in Japan, and quantitative reverse transcription polymerase chain reaction testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was performed in four airport quarantine stations (Narita, Hanada, Nagoya and Kansai airports; Supplementary Table S1). The Japan Ministry of Health, Labor and Welfare reported that there were 782 cases (of the 168 061 tests performed) of SARS-CoV-2 infection confirmed at airport quarantine stations between March and 1 September 2020. The SARS-CoV-2-positive RNA samples were subjected to whole genome sequencing based on a modified ARTIC Network protocol,1 and 129 full genome sequences were identified (Supplementary Table S2). A haplotype network analysis using genome-wide single-nucleotide variation was performed,2 including the near full-length complete genome sequences (≥29 kb; 9493 entries; Supplementary Table S3) retrieved from the Global Initiative on Sharing All Influenza Data (GISAID) EpiCoV database on 10 September 2020 (Figure 1).
Figure 1.
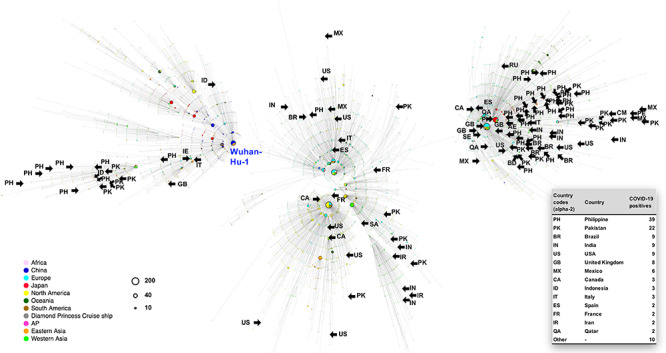
Haplotype network analysis using genome-wide single-nucleotide variations of worldwide SARS-CoV-2 isolates. Whole-genome sequences of SARS-CoV-2 isolates from COVID-19 positive patients at four international airport quarantines in Japan (n = 129) were compared, including all GISAID-available SARS-CoV-2 genomes (n = 9493, updated on 10 September 2020), using network analysis. A bold arrow with the country code (alpha-2) indicates the node of genome sequence, which was obtained from the patient coming from the most recent port of call in country. A country-specific highlighting images were presented in Supplementary Figure S1. COVID-19, coronavirus disease; GISAID, Global Initiative on Sharing All Influenza Data; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
Of the 129 genome sequences identified (Supplementary Table S2), 39, 22 and 9 genome sequences were identified primarily in travelers from the Philippines (PH), Pakistan (PK) and Brazil (BR), respectively (Supplementary Figure S1). As of September 1, PH, PK and BR were experiencing high-intensity COVID-19 epidemics, with 224 264, 296 149 and 3.9-million cumulative cases, respectively. As immigration restrictions began in Japan on 27 March 2020, there have been no outbreaks due to imported cases identified by the domestic genome surveillance program. Most recent SARS-CoV-2 isolates were progeny of the Japanese isolates of European origin (B.1.1 Pangolin lineage).2 However, attention should be paid to the potential risk of COVID-19 importation. Genome sequences originating from the quarantine stations may support international cooperation by providing information regarding country-related SARS-CoV-2 isolates, because of the limited number of genome sequence entries from these countries in GISAID (PH: 29, PK: 20).
The daily number of airport checks for COVID-19 would increase to ~20 000 following the recent relaxation of entry restrictions, and the partial reopening of Japan (on 1 October 2020) to foreign nationals (mainly business travelers) planning to stay in Japan for at least 3 months. Interestingly, there is a potential risk of COVID-19 during air travel.3 Being in close proximity to strangers with an unknown infection status for hours in an aircraft is a matter of concern, although physical distancing and use of face coverings or masks can reduce the risk during boarding.4 The number of foreigners entering Japan remains to be determined, but the resumption of foreign air travel could trigger a resurgence of COVID-19.
In conclusion, evaluation of imported SARS-CoV-2 genome sequences identified at quarantine stations in Japan until 31 August 2020, and haplotype network analysis demonstrated multiple genome lineages. To prevent the introduction of COVID-19, it is necessary to formulate an efficient strategy of testing and real-time genome sequencing to support quarantine monitoring based on the sentinel surveillance in travel medicine.5
Ethical statement
The study protocol was approved by National Institute of Infectious Diseases in Japan (Approval No. 1091). The ethical committee waived the need for written consent regarding research into the viral genome sequence.
Data availability statement
The new sequences have been deposited in the Global Initiative on Sharing All Influenza Data (GISAID) with accession IDs (Supplementary Table S2).
Authors’ contributions
TS, KI, MO, TW and MK designed and organized the genome study. RT and MH performed genome sequencing. TS, KI, KY and TKS performed the genome analysis. MK wrote the manuscript.
Supplementary Material
Acknowledgements
We are grateful to the staff of airport quarantines (Narita, Hanada, Nagoya and Kansai airports) and the Ministry of Health, Labor and Welfare for their assistance with field investigations, data collection, laboratory testing, and collection and transportation of clinical specimens. We would like to thank all researchers who kindly deposited and shared genomic data on GISAID. The genome sequence acknowledgments are presented in Supplementary Table S3. This work was supported by a Grant-in-Aid from the Japan Agency for Medical Research and Development Research Program on Emerging and Re-emerging Infectious Diseases [JP19fk0108103 and JP19fk0108104, respectively].
Conflict of interest: None declared.
Contributor Information
Tsuyoshi Sekizuka, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Kentaro Itokawa, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Koji Yatsu, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Rina Tanaka, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Masanori Hashino, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Tetsuro Kawano-Sugaya, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Makoto Ohnishi, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Takaji Wakita, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
Makoto Kuroda, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan.
References
- 1. Itokawa K, Sekizuka T, Hashino M et al. Disentangling primer interactions improves sars-cov-2 genome sequencing by multiplex tiling PCR. PLoS One 2020; 15:e0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sekizuka T, Itokawa K, Hashino M et al. A genome epidemiological study of sars-cov-2 introduction into Japan. mSphere 2020; 5. doi: 10.1101/2020.07.01.20143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pombal R, Hosegood I, Powell D. Risk of covid-19 during air travel. JAMA 2020; 324:1798. [DOI] [PubMed] [Google Scholar]
- 4. Khanh NC, Thai PQ, Quach HL et al. Transmission of severe acute respiratory syndrome coronavirus 2 during long flight. Emerg Infect Dis 2020; 26. doi: 10.3201/eid2611.203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilder-Smith A, Boggild AK. Sentinel surveillance in travel medicine: 20 years of geosentinel publications (1999-2018). J Travel Med 2018; 25. doi: 10.1093/jtm/tay139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The new sequences have been deposited in the Global Initiative on Sharing All Influenza Data (GISAID) with accession IDs (Supplementary Table S2).


