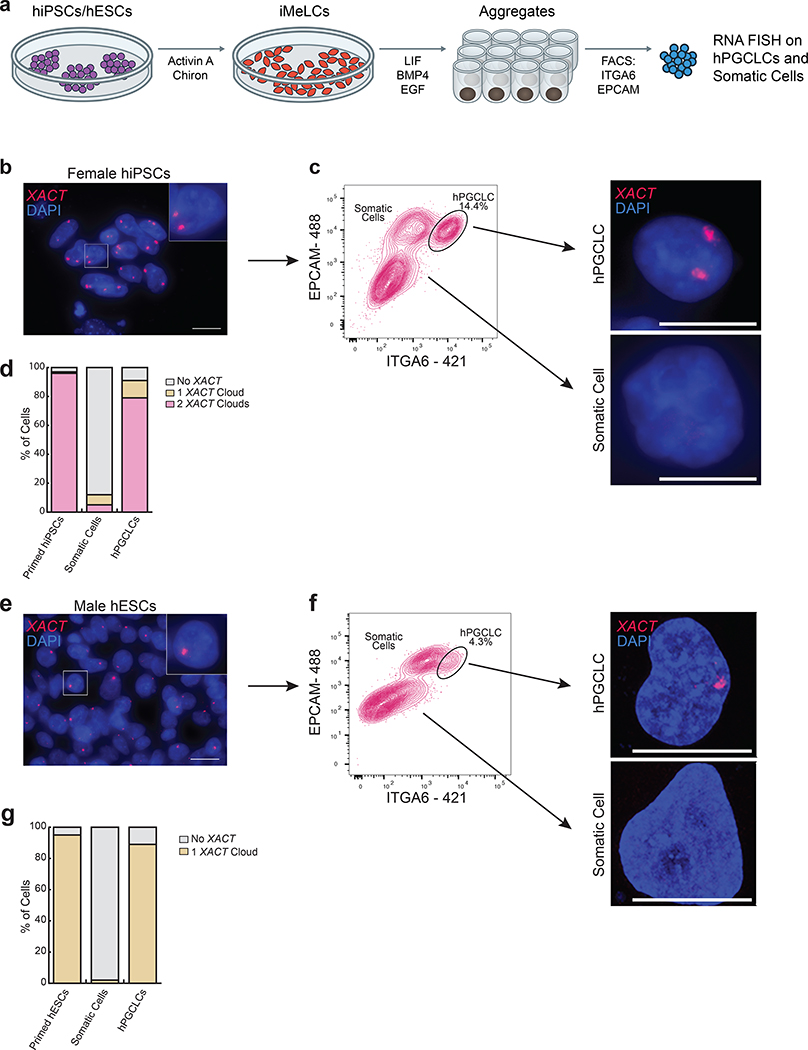
Fig. 2. The lncRNA XACT is restricted to male and female hPGCLCs and is not expressed in somatic cells in vitro.
a. Differentiation of hPGCLCs from hiPSCs or hESCs through a incipient mesoderm like cells (iMeLC) intermediate. PGCLCs and somatic cells within the aggregates are separated at day 4 (D4) by FACS using antibodies that recognize EPCAM and ITGA6. b. RNA FISH for XACT in primed female hiPSCs (MZT04-J), that harbor a Xa and an eroded X-chromosome (Xe) with XACT (red) and DAPI (blue) stainings. XACT clouds are detected from the Xa and Xe; scale bar 20 microns. The experiments have been performed twice with similar results. c. Female hiPSCs are differentiated to hPGCLCs and isolated from the aggregates by FACS at D4. The hPGCLC population is indicated. Right panel: XACT RNA FISH in hPGCLCs and somatic cells. The experiments have been performed twice with similar results; scale bar 10microns. d. Quantification of the proportion of cells with different numbers of XACT clouds in starting hiPSCs (n=100 cells), hPGCLCs (n=82 cells) and somatic cells (n=100 cells) from (c). e. XACT RNA FISH (red) in male hESCs (UCLA2); scale bar 20 microns similar to (b). f. As in (c), except for UCLA2 hESCs; the experiments have been performed twice with similar results, scale bar 10microns. g. Quantification of the proportion of cells with a different number of XACT clouds in UCLA2 hESCs (n=100 cells) and derived hPGCLCs (n=92 cells) and somatic cells (n=100 cells pooled from the two experiments). Statistical source data are provided in Source data fig. 2.