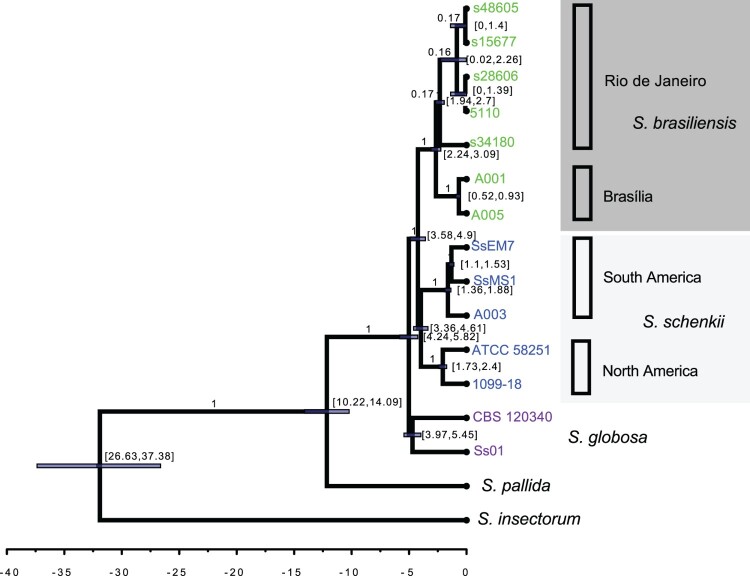
Figure 1.
Whole-genome Sporothrix genealogy using BEAST analyses. The phylogenomic tree was inferred using Bayesian inference under the GTR model with gamma variation among sites and S. insectorum was set as the outgroup. The 90% confidence interval marking the separation between S. brasiliensis and S. schenckii was set to 3.8 - 4.9 MYA, a rate interval of [0.9E-3, 16.7E-3] based on evolutionary rate estimation based on different groups of fungi and a birth-death tree prior with incomplete sampling were set as priors. The clade distribution, posterior probabilities and divergence times are shown.