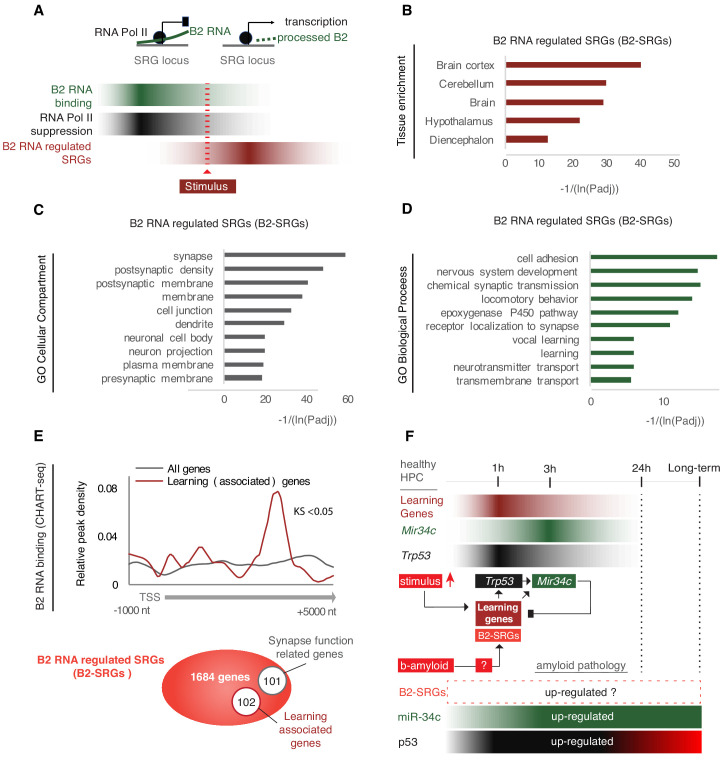
Figure 1. B2-SRGs are enriched in neural functions.
(A) Regulation mode of SRGs by B2 RNA processing based on previous works (Zovoilis et al., 2016; Yakovchuk et al., 2009; Ponicsan et al., 2010). Color intensity represents higher B2 RNA binding (green), Pol II suppression (black) or SRG expression (red). (B) Tissue enrichment analysis of B2-SRGs listed in Supplementary file 1 based on the DAVID functional annotation platform (Huang et al., 2009a; Huang et al., 2009b). The adjusted p-values of top five ranking terms are plotted as a function with higher scores representing lower adjusted p-values (higher statistical significance) ranging from padj 1.92E-04 for ‘Diencephalon’ to padj 8.91E-13 for ‘Brain Cortex’. A complete list of all terms that pass the EASE 0.05 score threshold is presented in Supplementary file 1. (C) Gene ontology (GO) analysis (DAVID) of B2 RNA regulated genes on the basis of cellular compartments. The adjusted p-values of top 10 ranking terms are plotted as a function with higher scores representing lower adjusted p-values ranging from 2.23E-06 for ‘Presynaptic membrane’ to 1.88E-18 for ‘Synapse’. A complete list of all terms that pass the EASE 0.05 score threshold is presented in Supplementary file 1. (D) GO analysis (DAVID) of B2 RNA regulated genes on the basis of biological processes. As above, adjusted p-values of top ranking terms are plotted as a function, with higher scores representing lower adjusted p-values ranging from 0.02 for ‘Transmembrane transport’ to 5.44E-06 for ‘Cell adhesion’. A complete list of all terms that pass the EASE 0.05 score threshold is presented in Supplementary file 1. (E) Upper panel: Metagene analysis of distribution of genomic B2 RNA-binding sites across the start of genes from Zovoilis et al., 2016, comparing learning-associated genes from Peleg et al., 2010 with all genes (Kolmogorov Smirnov test, KS <0.05). TSS represents the Transcription Start Site of these genes. Lower panel: Representation of the number of B2-SRGs that are (i) learning associated based on Peleg et al., 2010 (Supplementary file 1) and (ii) synapse function related based on GO term enrichment (Supplementary file 1, right panel). (F) Regulatory loop of learning genes-Trp53-Mir34c in mouse hippocampus based on Peleg et al., 2010; Zovoilis et al., 2011; Yamakuchi and Lowenstein, 2009. A potential role of B2-SRGs, that include many learning genes, in affecting this loop in amyloid pathology remained unknown after these studies (noted with a question mark in the figure).