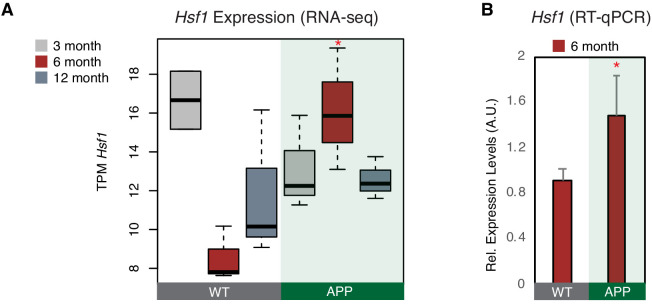
Figure 4. Hsf1 is upregulated in 6-month-old APP mice.
(A) Boxplot depicts distribution of expression levels of Hsf1 gene among different age groups of mice between wild type and APP. Values are based on TPM counts of long-RNA-seq data. Statistical significance (p value threshold 0.05) for the comparison between 6-month-old APP and 6-month-old WT (p=0.03) (n = 3/group, unpaired non-directional t-test). All the other comparisons between groups and different ages are non-significant. (B) Expression levels of Hsf1 in 6-month-old mice as defined by RT-qPCR. Statistical significance (p value threshold 0.05) for 6-month-old APP greater than 6-month-old WT (depicted as asterisk, unpaired directional t-test, n = 3/group, error bars represent standard deviation from the mean).