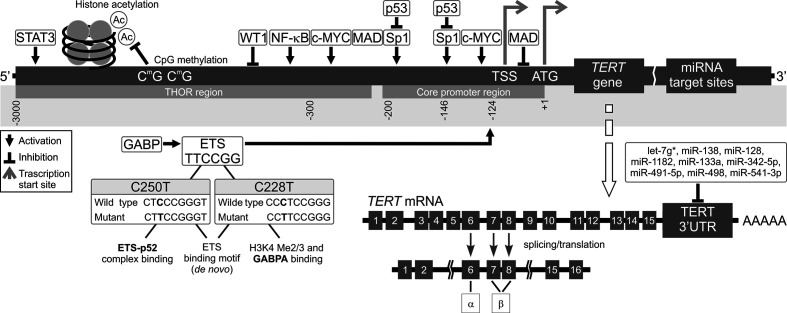
Figure 2.
Mechanisms of TERT transcription regulation. The figure shows various mechanisms regulating TERT expression at the transcriptional level. Transcription factors: activators (e.g., c-MYC, SP1, STAT3, NF-κB, and ETS), repressors (e.g., MAD, p53, and WT1), and their respective binding sites are shown. Binding of these transcriptional agents to TERT could be controlled by DNA methylation (CpG sites) in the TERT Hypermethylation Oncological Region (THOR). Two main hotspot mutations within TERTp, -146C > T (C250T) and -124C > T (C228T) upstream of the transcription start site (TSS) generate new E-twenty-six (ETS) binding sites, leading to GABP recruitment and, eventually, TERT transcription. Alternatively spliced variants of TERT, which do not have telomerase activity, could be also generated. Most tissues and organs express no or very low levels of TERT mRNA, dependent on histone markers that are correlated with passive or active transcription in many cells. The figure also shows different miRNAs at the 3’UTR that inhibit translation of TERT.