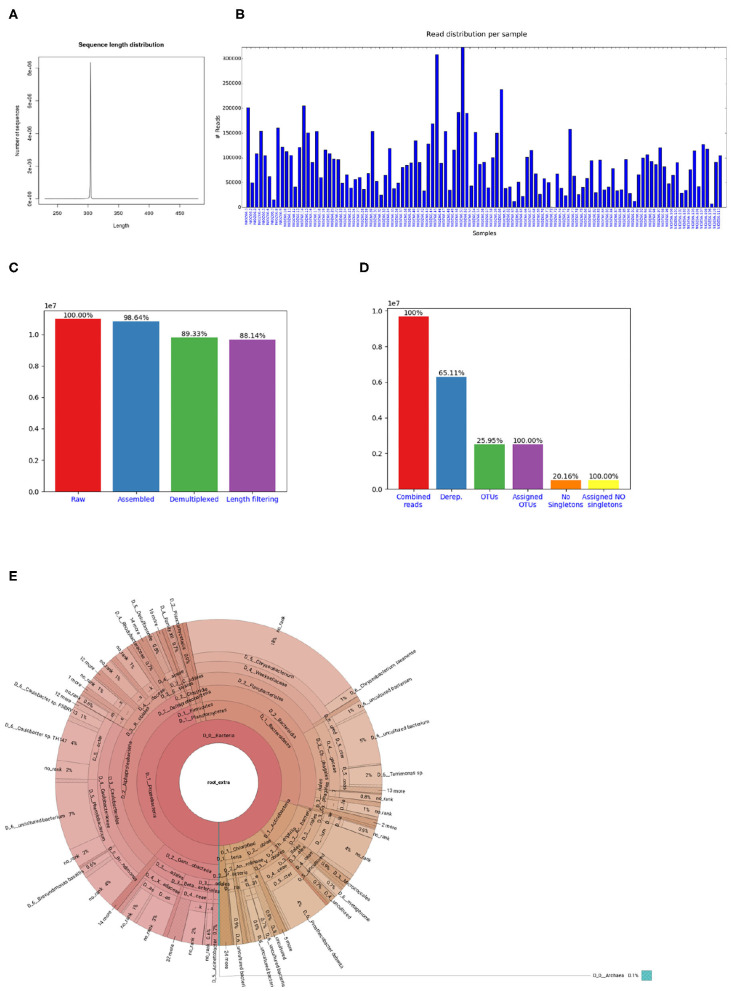
Figure 3.
Figures shown in Cascabel reports. (A) Smoothed sequence length distribution after merging reads, for one library. The plot is meant to help making a sensible choice for sequence length filtering. (B) Number of sequences per sample. This histogram is part of the OTU report (including all libraries). (C) Number of sequences after individual pre-processing steps. “Assembled” refers to the number of raw read pairs which could be merged based on their overlap. “Demultiplexed” refers to the number of raw reads which could be assembled and assigned to a sample, and “Length filtering” indicates the number of raw reads passing the previous and the sequence length criteria. This plot is part of the library report. (D) Number of sequences after individual steps after potentially combining several libraries (total number of reads) and generating OTUs. “Derep.” indicates the number of dereplicated reads and their percentage relative to the total combined reads. “OTUs” is the total number of OTUs and the percentage is relative to the number of combined reads. “Assigned OTUs” is the number and percentage of OTUs with a taxonomic assignment. “No singletons” refers to the number and percentage of OTUs excluding singleton OTUs, and “Assigned NO singletons” is the number and percentage of singleton-free OTUs with a taxonomic assignation. The plot is part of the OTU report. (E) Krona chart for one sample. The krona charts are interactive and can be viewed with a web browser. Colors indicate the taxonomic groups to which the OTU was assigned. Each ring of the pie chart represents a different taxonomic level. An example of a full library report is shown in Supplementary Datasheet 3, and an OTU report is provided in Supplementary Datasheet 4.