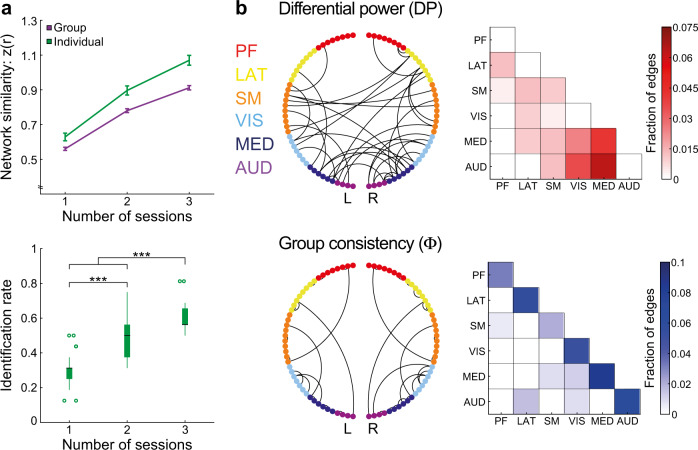
Fig. 2. Factors contributing to characterization of individual variation.
a Network similarity values (top) and identification rates (bottom) as a function of number of included sessions in the two connectivity matrices of each mouse. All combinations of data sampling of one, two or three sessions were calculated and compared using repeated-measures ANOVA (network similarity, see text) or two-tailed unpaired student t-test (identification rate, ***P < 0.001). Error bars (top) represent the standard error of the mean (n = 16 mice); boxplots (bottom) represent the median (center line), interquartile range (box limits); 1.5 × interquartile range (whiskers) and outlier (points), n = 30, n = 90, n = 40 rates derived from connectome-based identification analyses of all combinations of one, two, and three session per half of data, respectively. b Unique (DP, top) and consistent (Φ, bottom) edges in individual connectomes. For visualization purposes, both measures were thresholded at the 99th percentile. In the circle plot (left) the 86 nodes are organized based on their anatomical module identity and cortical hemisphere; lines indicate edges. In the matrices (right), the fraction of edges connecting between and within modules is color coded, with darkly shaded cells representing higher DP (top) and Φ (bottom) values. PF, Prefrontal; LAT, Lateral; SM, Somatomotor; VIS, Visual; MED, Medial; AUD, Auditory.