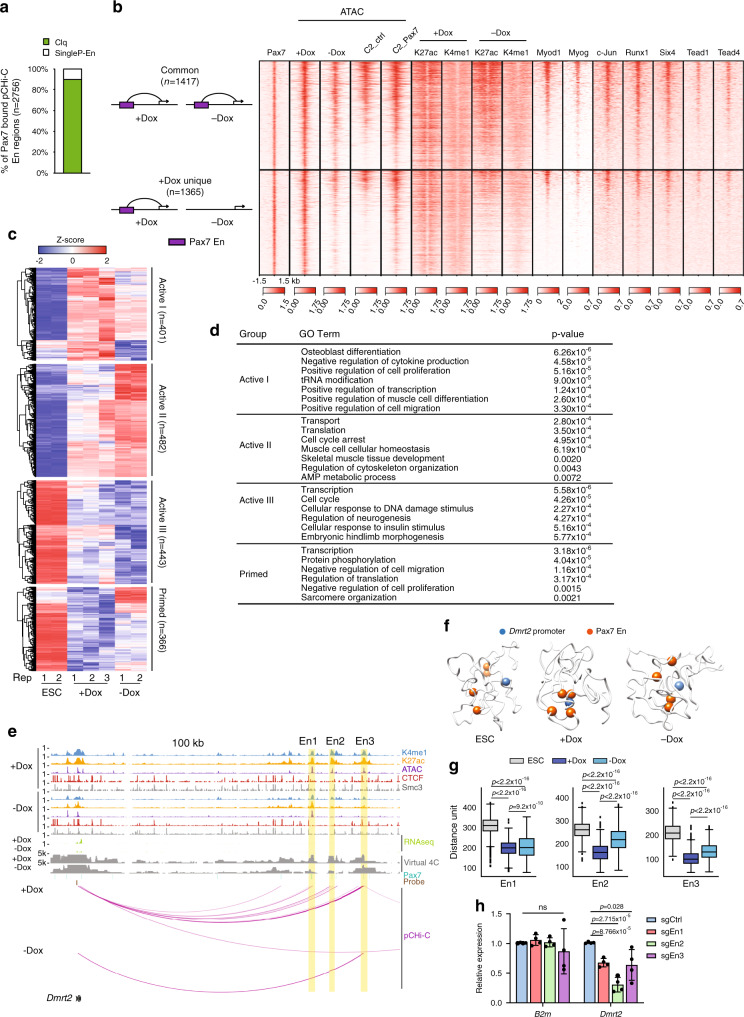
Fig. 4. Two types of Pax7 enhancers collaborate to activate or prime transcription.
a Quantification of Pax7 enhancers involved in P–En cliques (Clq) in +Dox iPax7 cells (n = 2756). b Promoter-interacting Pax7 enhancers show two distinct patterns of accessibility, histone modifications, and transcription factor binding. (Left) Schematic representation of two types of promoter-Pax7 enhancer (En) interactions. (Right) Heat map of ChIP-seq (TFs and histone modifications) and ATAC-seq signals centered on Pax7 sites (±1.5 kb) suggests preferential co-localization of myogenic transcription factors at Pax7 enhancers involved in common promoter-Pax7 enhancer interactions. C2_ctrl, control C2C12 cells expressing 3xFlag; C2_Pax7, C2C12 cells expressing Pax7_3xFlag. c Classification of differentially transcribed genes with Pax7 enhancer interactions detected by pCHi-C, according to their expression levels in ESC and iPax7 populations, as shown. Differentially expressed genes were selected using one-way ANOVA (cut-off padj of 0.05). Normalized RNA-seq data for each sample were used to calculate z-scores. Genes were grouped into four clusters based on the pattern of average z-scores across the three cell populations, and each group was further classified with hierarchical clustering. d GO analysis for the four groups of genes from c. e Genome browser tracks around Dmrt2 locus, showing ChIP-seq, ATAC-seq, RNA-seq, Pax7 binding sites, pCHi-C probes, and magenta arcs representing high-confidence pCHi-C interactions (with corresponding virtual 4C plots) in iPax7 cells before (+Dox) and after (−Dox) differentiation. Yellow, Pax7 enhancer regions (En1–3) that loop to the Dmrt2 promoter. f 3D chromatin conformation models for Dmrt2 locus based on pCHi-C data. The top-scoring 3D models for Dmrt2 in ESCs and iPax7 cells are shown. These models show that the three Pax7 enhancers (En1–3) tend to position closer to the Dmrt2 promoter in Dox-treated iPax7 cells. g Distance distributions between the Dmrt2 promoter and three Pax7-bound enhancers (En1–3), highlighted in f, in ESCs and iPax7 cells, obtained from the ensemble of models (n = 975) built with the data from each of the cell lines. The boxes denote the 25th and 75th percentile (bottom and top of box), and median value (horizontal band inside box). The whiskers indicate the values observed within up to 1.5 times the interquartile range above and below the box. Statistical significance tested with Kolmogorov–Smirnov test. h Relative expression of Dmrt2 and B2m (β2 microglobulin) in iPax7 progenitors after CRISPR inhibition with sgRNAs targeting Pax7 sites within Dmrt2 En1–3; n = 4 (biological replicates for each sgRNA). Bars are mean ± s.d., normalized by Gapdh and expressed relative to mean levels of the control sgRNA (sgCtrl). Statistical significance tested with two-tailed Student’s t-test.