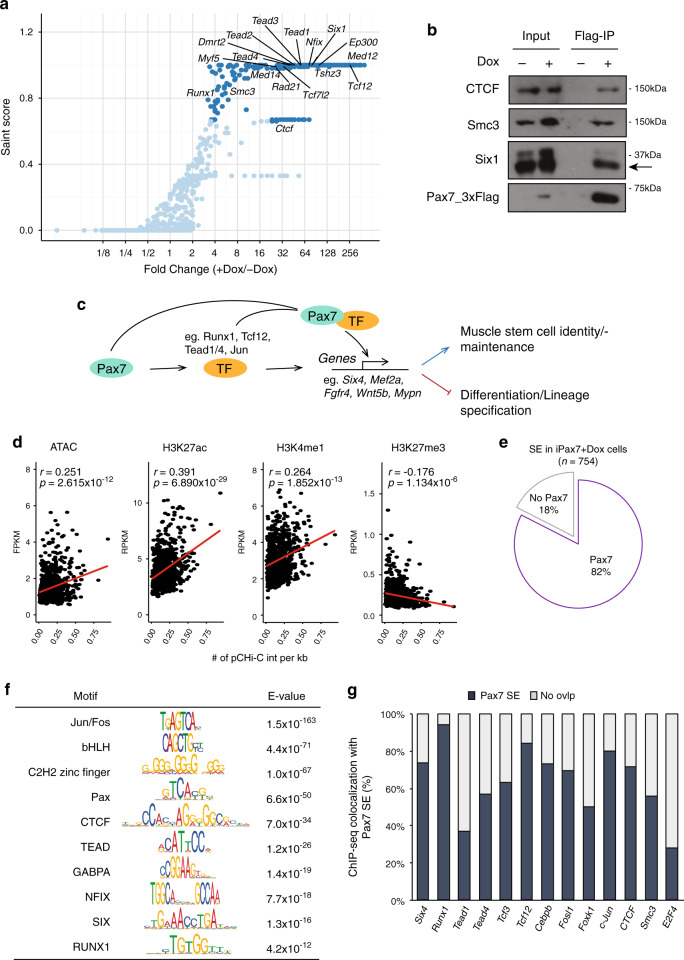
Fig. 5. Pax7 assembles feed-forward loops with its target TFs.
a SAINT analysis revealing high-scoring Pax7-associated proteins detected by IP-MS. Proteins identified as high-confidence interactors of Flag-tagged Pax7 are marked in dark blue; n = 3 (biological replicates). b Immuno-blot confirming interactions between indicated proteins and Flag-tagged Pax7 in iPax7 cells. c Schematic representation of a feed-forward regulatory loop assembled by Pax7 and its target TFs. d Pairwise Pearson correlation between the number of high-confidence pCHi-C interactions per kb and signals for ATAC-seq and ChIP-seq for histone modifications at SEs in iPax7 muscle progenitors. A linear regression line (y ~ x) is plotted in red; r, Pearson correlation coefficient; p values determined by two-tailed Student’s t-test. e Venn diagram indicates that most SEs identified in +Dox iPax7 cells that display high-confidence pCHi-C interactions contain Pax7-bound active enhancers. f Highly enriched TF binding motifs identified in SEs with pCHi-C interactions in +Dox iPax7 cells. The E-value is the p value derived from a one-tailed binomial test multiplied by the number of motifs in the input database. g Quantification of ChIP-seq peaks detected at Pax7 site-containing SEs. E2F4, control TF not detected as a Pax7 interactor.