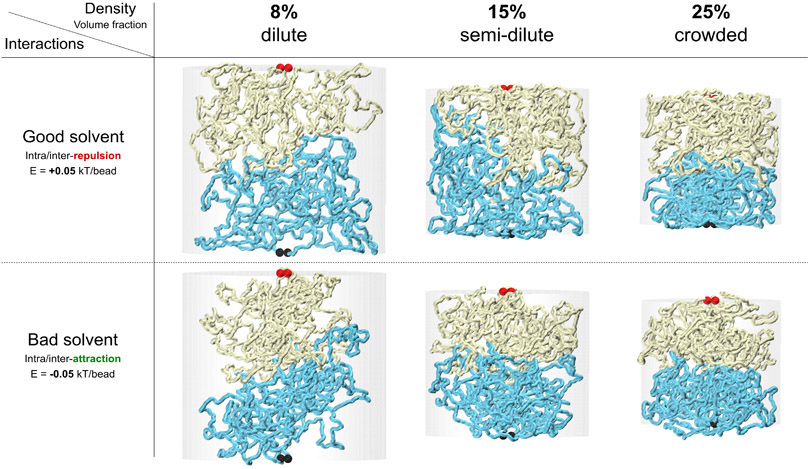
Extended Data Fig. 5. Simulation snapshots for domain interfaces at different solution conditions.
Equilibrated simulation snapshots for domain interfaces at different solution conditions. Two major determinants of polymer mixing are (1) whether polymer prefers to interact with the solvent (good solvent) or to interact with itself (bad solvent). This can be simulated effectively by a repulsive/attractive short-range interaction between the chain’s beads. Both conditions might be present in the nucleus leading to microphase separation of chromatin into euchromatin versus heterochromatin domains; and (2) the density of the polymer mixture, here simulated by the confining cylindrical walls surrounding the two chains. Our simulations at different solvent and under different crowding conditions still show that domains that are topologically unlinked will barely mix and retain their smooth interfaces. The result might be different for the good solvent in ultra-dilute condition, in which, domains will barely touch and won’t generate measurable multi-contact walks. We believe this condition is not relevant to the chromatin in vivo and would not be consistent with our MC-3C data.