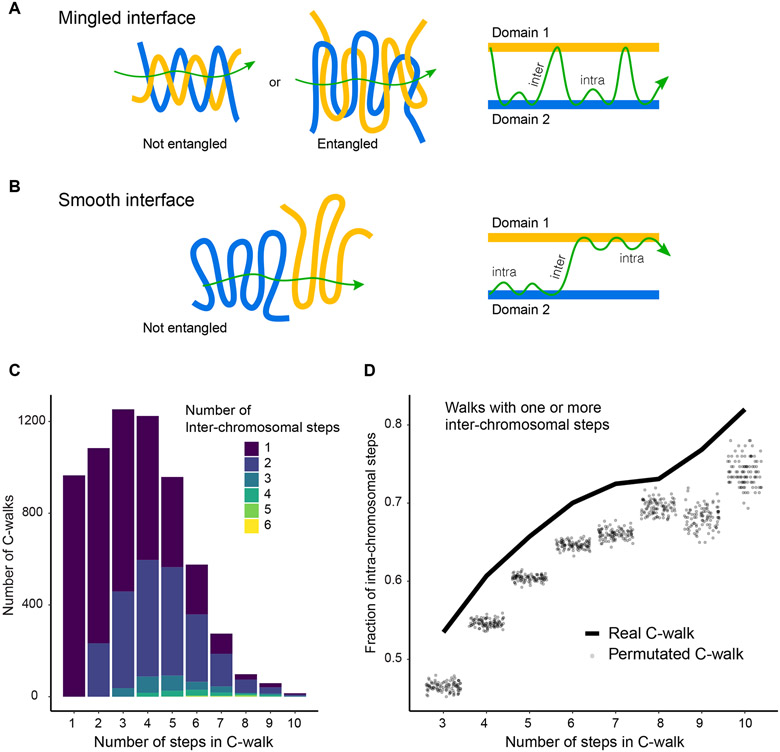
Figure 2:
Inter-chromosomal interfaces show limited mixing. A, B: Schematic of how mingled and smooth interfaces would be captured in multi-contact data. In a mingled interface (top row) a C-walk would contain many steps between the two domains, as schematically indicated on the right. In a smooth interface the C-walk would contain series of steps within one domain and only occasionally contain a step between the two. Here, domains are any two distinct chromosomes or two distal A and/or B compartments located on the same chromosome. In general, any two chromatin regions which are not neighbors along the linear genome but are spatially proximal would qualify as (interacting) domains in this picture. C: Stacked barplots with the number of C-walks containing at least one inter-chromosomal step separated by the number of steps between two chromosomes (in colors) and the number of steps (along the x-axis) that they contain. Most inter-chromosomal C-walks contain only one or two inter-chromosomal step. D: Proportion of intra-chromosomal steps in the set of C-walks with one or more inter-chromosomal steps, separated by the total number of steps in each C-walk. The solid line shows the proportion for real C-walks. The dots correspond to the same proportion in 100 sets of permutated walks. The observed C-walks have a higher proportion of intra-chromosomal steps than would expected if the order of the steps was random. Sum of p-values is less than 10−7. Source data for panels c and d area available online.