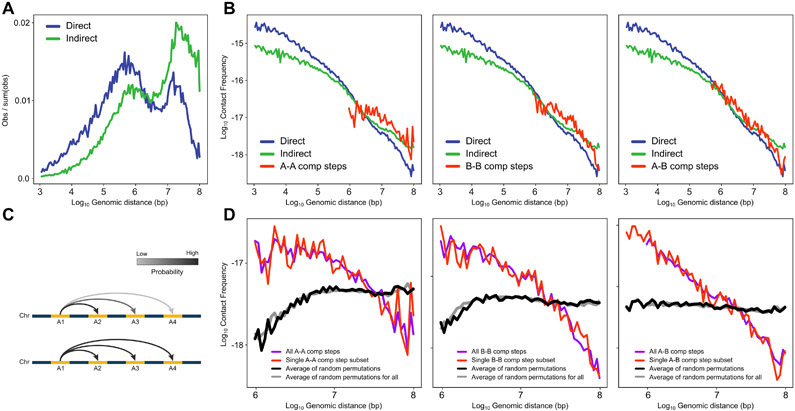
Extended Data Fig. 3. Ruling out the impact of random ligation events.
Frequency of intra- and inter-domain interactions as a function of the genomic distance. Most long-range interactions are inter-domain steps. We considered the possibility that the inter-domain steps are experimental noise due to random ligation events that do occur with some frequency in 3C-based experiments. To investigate this, here, we determined whether inter-domain steps display the expected inverse relationship with genomic distance that separates the loci. We find that the frequency of inter-domain steps decays rapidly. Such distance-dependent decay would not be expected if these interactions were the result of random ligation. A. Density of interactions corresponding to Figure 1D with logarithmic binning similar to Bonev et al. (54). This graph shows clearly that indirect interactions are more long range. B. Contact probability profiles for steps in walks with single inter-compartment step (A-A, B-B, A-B). These steps follow a similar distance decay as direct and indirect interactions. This shows that inter-compartment steps are not random ligation events. C. Schematic of inter-domain interactions with (upper) or without (lower) distance decay. D. Observed distance decay scaling for C-walks with inter-compartment steps and the permutated versions of those walks. Left Panel (A-A): All inter-compartment A-A steps (purple; 2,980 interactions from all C-walks that contain one or more inter-compartment steps) and single inter-compartment A-A (red; 936 interactions from C-walks that contain only a single inter-compartment step). Middle Panel (B-B): All B-B steps (purple; 2,720 interactions) and single B-B (red; 882 interactions). Right Panel (A-B): All A-B steps (purple; 5,655 interactions) and single A-B (red; 1,392 interactions). The permutated walks mimic the effect of random ligation between distal domains. Randomized walks generate flat scaling profiles not consistent with the observations.