Fig. 6. Downregulation of miR-106a~363 cluster of miRNAs (including miR-363–3p, miR-20b-5p and miR-106a-5p) drives NEPC via pleiotropic regulation of multiple NEPC drivers.
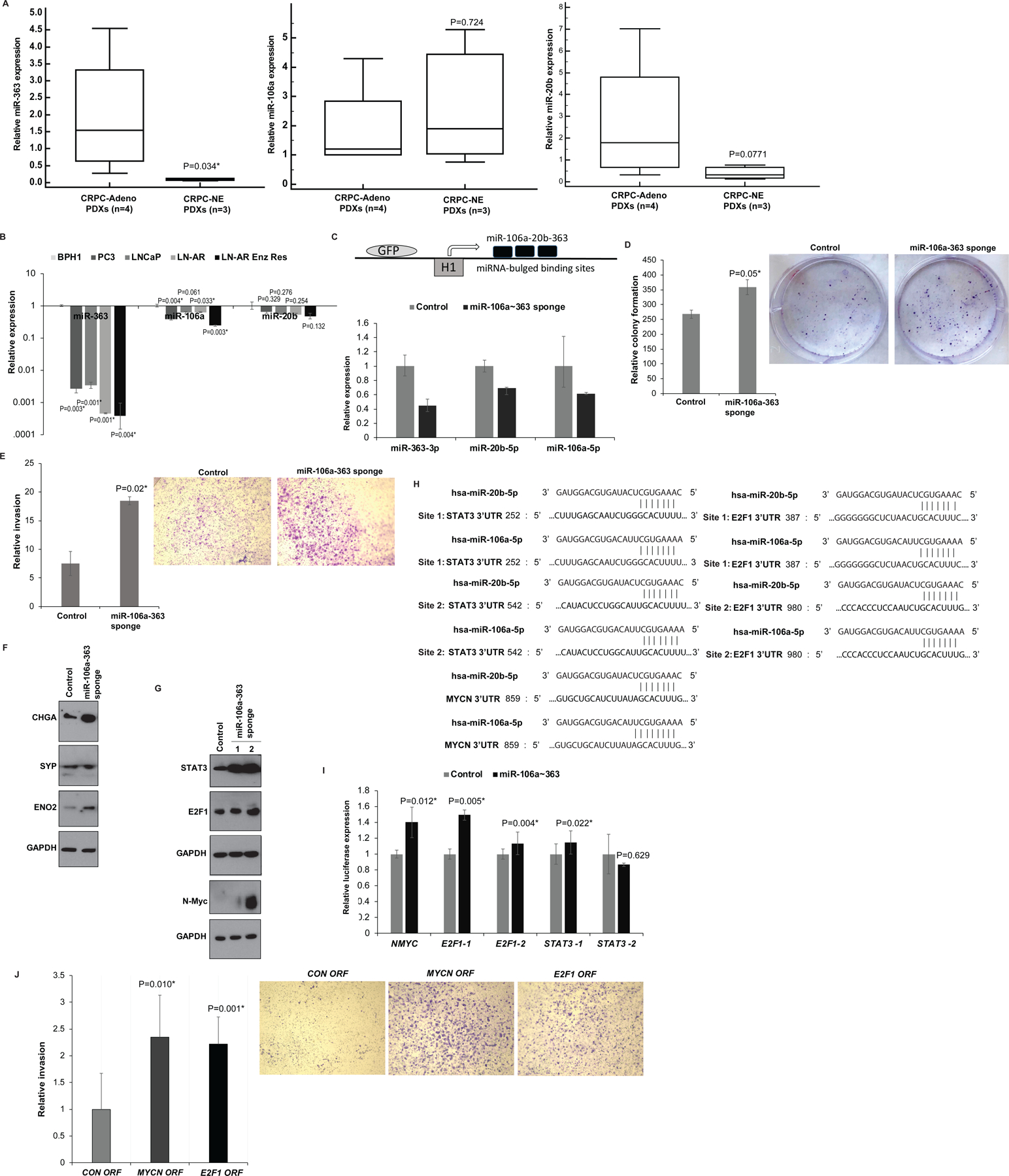
A. Real time PCR analyses of miR-363 (left panel), miR-106a (middle panel) and miR-20b (right panel) in CRPC-NE (LuCaP 49, 145.1 and 145.2) vs CRPC-Adeno PDX models (LuCaP 70, 78, 81 and 92). RNU48 was used as an endogenous control.
B. Real time PCR analyses of miR-363 (left panel), miR-106a (middle panel) and miR-20b (right panel) in benign prostatic hyperplasia cell line (BPH1) and PCa cell lines (PC3, LNCaP, LNCaP-AR, LNCaP-AR-Enzalutamide resistant). RNU48 was used as an endogenous control.
C. Upper panel: Schematic representation of miR-106a~363 sponge construct used for miR-106a~363 downregulation. This construct includes an engineered RNA molecule with multiple bulged miR binding sites, driven by lentiviral pGreen expression system that employs the RNA polymerase III-driven H1 promoter. Lower panel: Real time PCR based expression of miR-106a, miR-363 and miR-20b after stable transduction of control/miR-106a~363 targeting sponge construct in C42B cells.
D. Relative colony formation ability upon control/miR-106a~363 sponge treated-stable C42B cells.
E. In vitro invasion assay in control/ miR-106a~363 sponge-transduced C42B cells.
F. Western blot analyses for CHGA, SYP and ENO2 in control/miR-106a~363 sponge-transduced C42B cells. GAPDH was used as a loading control.
G. Western blot analyses for indicated proteins in control/miR-106a~363 sponge-transduced C42B cells. GAPDH was used as a loading control.
H. Schematic representation of STAT3, E2F1 and MYCN 3’ UTR regions showing potential miR-106a and miR-20b binding sites.
I. Luciferase reporter assays with control 3’ UTR construct or STAT3/E2F1/MYCN 3’ UTR constructs in control/miR-106a~363 sponge-transduced C42B cells. Luciferase activities (Firefly/renilla) were calculated relative to corresponding control 3’UTR constructs and plotted as relative values. Error bars represent SD.
J. C42B cells were transiently transfected with control/ MYCN/E2F1 ORF constructs for 72 hours followed by in vitro transwell invasion assay.
