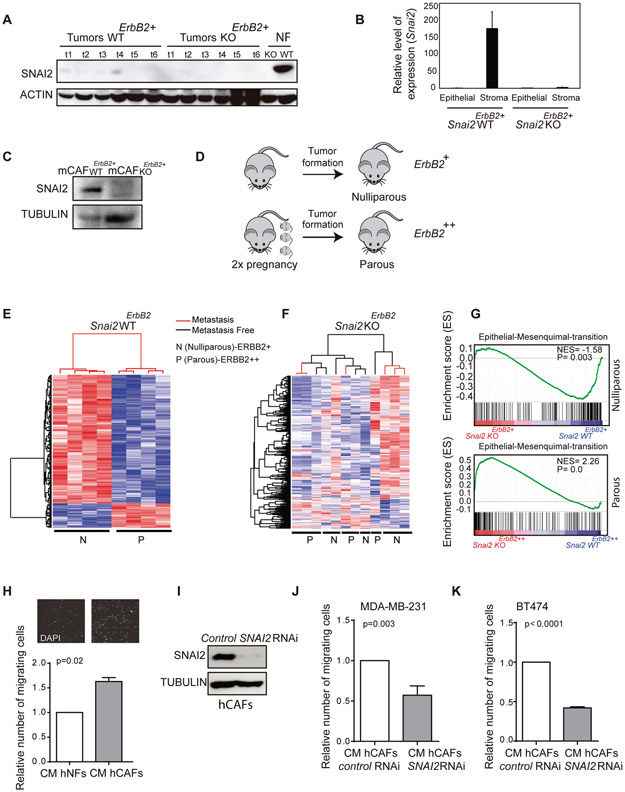
Fig. 2. The profile of breast tumor gene expression is affected by SNAI2 deficiency in a non-cell-autonomous manner.
(A) Detection of SNAI2 in Snai2 WTErbB2 and Snai2 KOErbB2 tumors by western blot. Mouse embryonic fibroblasts (MEFs) were used as a positive control for SNAI2 expression. (B) QPCR quantification of Snai2 in the epithelial and stromal components from Snai2 WTErbB2+ and Snai2 KOErbB2+ tumors. Snai2 RNA is not detected in Snai2 WTErbB2+ mice by QPCR after EpCAM+ cell sorting of the epithelial component, yet it is found in the stromal CD140+ cells. (C) SNAI2 protein in western blots of CAFs from mouse breast tumors. (D) Scheme representing the study design. Tumors from Snai2 WTErbB2 and Snai2 KOErbB2 mice were analyzed, both from nulliparous (ERBB2+ expression: WT N=41 and KO N=33) and parous (ERBB2++ expression) females in which the tumors developed after two pregnancies (WT, N=33, and KO, N=22). (E) The 602 gene expression signature allows unsupervised classification of the tumors from Snai2 WTErbB2 nulliparous and parous mice. (F) The signature does not permit the clustering of Snai2 KOErbB2 nulliparous and parous mice. (G) GSEA analysis: representation of the enrichment of the EMT pathway in tumors from Snai2 WTErbB2+ relative to the tumors from Snai2 KOErbB2+ nulliparous mice (above), and in tumors Snai2 KOErbB2++ relative to the tumors from Snai2 WTErbB2++ parous mice (below). (H) Conditioned medium (CM) from cultured human normal fibroblasts (hNFs) or human cancer-associated fibroblasts (hCAFs) was used to induce MDA-MB-231 cell migration (N=3). (I) Representative western blot of normal hCAFs and hCAFs in which SNAI2 was depleted by specific siRNAs, probed for SNAI2. (J) Migration of MDA-MB-231 cells in the presence of CM from control or SNAI2 depleted hCAFs (N=3). (K) Migration of BT474 cells using CM from control or SNAI2 depleted hCAFs (N=3). Statistical significance in panels I, K was evaluated using an unpaired t-test. In E and F red and blue color represent over-expressed and down-expressed genes, respectively.