Fig. 5 |. Sensitivity of ROSALIND reactions can be tuned using an RNA feedback circuit.
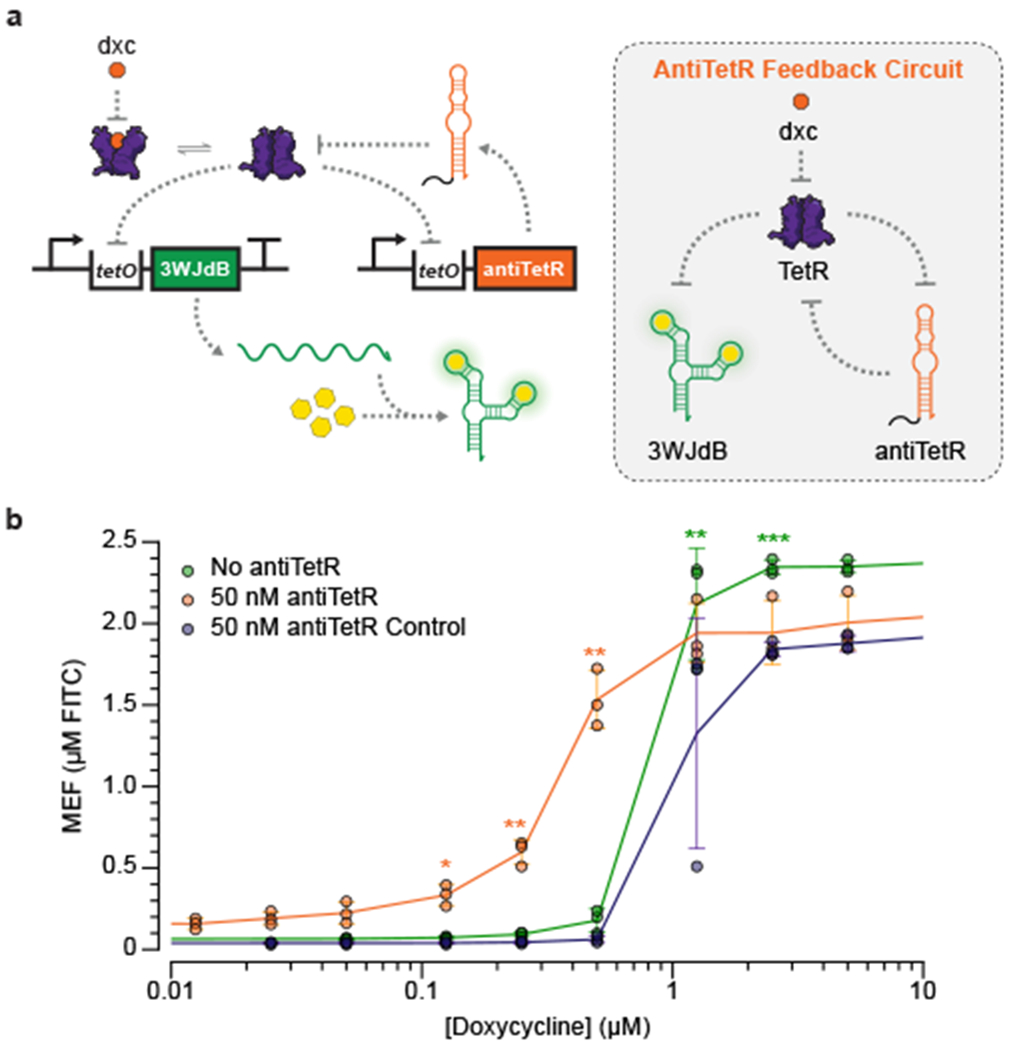
a, A TetR binding RNA, antiTetR, can be used to improve the limit of detection of the TetR sensor. In this feedback circuit, TetR regulates both the expression of 3WJdB and antiTetR. In the presence of the cognate ligand, doxycycline (dxc), antiTetR is transcribed and further de-represses TetR to enhance 3WJdB expression. b, When implemented in ROSALIND, the antiTetR feedback circuit improves the sensitivity of TetR by an order of magnitude. Each reaction contains 25 nM of the 3WJdB template, 50 nM of the antiTetR template and 1.25 µM of TetR dimer. A transcription template encoding the reverse sequence of antiTetR was used as a control to demonstrate that sensitization required the antiTetR interaction. All data shown for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC). Error bars indicate the average value of 3 independent biological replicates ± standard deviation. The ligand concentrations at which the signal is distinguishable from the background were determined using two-tailed heteroscedastic t-test against the no ligand condition, and their p-value ranges are indicated with asterisks (*** < 0.001, ** = 0.001 – 0.01, * = 0.01 – 0.05). Exact p-values along with degrees of freedom can be found in Supplementary Data File 3. Data for no ligand condition were excluded in b since the x-axis is on the log scale and are presented in Supplementary Data File 3.
