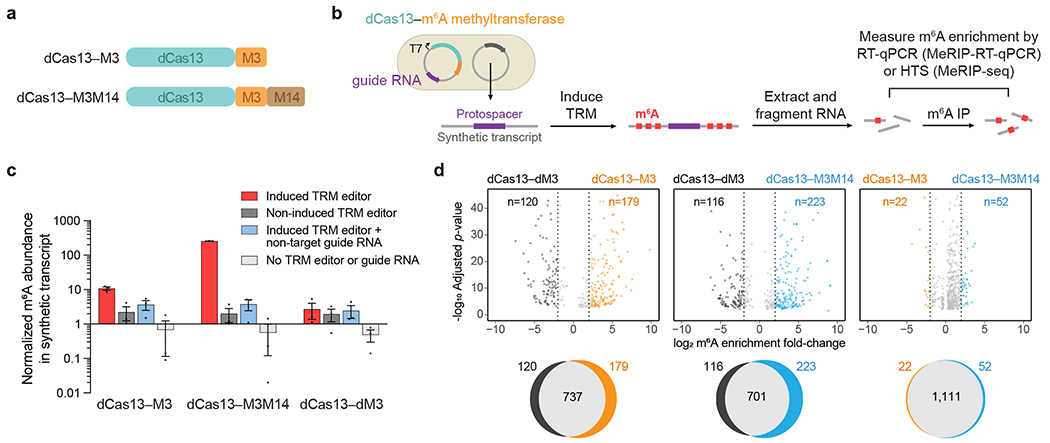
Figure 2. Validation of targeted RNA methylation (TRM) in E. coli.
(a) METTL3273-580 (M3) or METTL3359-580–(GGS)10–METTL14111-456 (M3M14) methyltransferases were fused to catalytically-impaired PspCas13b Δ984-1090 (dCas13) to generate dCas13–M3 or dCas13–M3M14 TRM editors. (b) Plasmids transformed into E. coli encode IPTG-inducible TRM editors with a guide RNA targeting a synthetic transcript. After targeted methylation, cellular total RNA was purified, fragmented, and immunoprecipitated with anti-m6A antibodies. Enrichment of m6A was measured by target-specific RT-qPCR (MeRIP-RT-qPCR) or transcriptome-wide high-throughput sequencing (MeRIP-seq). (c) Target transcript m6A abundance measured by MeRIP-RT-qPCR under four conditions: induced TRM editor with transcript-targeting guide, non-induced editor with transcript-targeting guide, induced editor with a non-target guide, and cells lacking TRM editor and guide plasmid. The methyltransferase-inactive control was dCas13–M3D395A (dCas13–dM3). Values and error bars reflect the mean±s.e.m. of n=3 independent biological replicates. (d) Top: differential m6A enrichment of all methylated adenines in E. coli expressing synthetic transcript-targeting guide RNAs and the indicated TRM editors. For the comparisons above, only differentially methylated sites with statistical significance (P<0.001) are shown. Bottom: Venn diagrams depicting overlap of all methylated m6A sites for the above comparisons. 42,418 total m6A motifs (RRACH) are susceptible to modification within the E. coli transcriptome. MeRIP-seq analysis was performed with n=3 independent biological replicates. Statistical significance was calculated using a logs likelihood-ratio test with false discovery rate correction.